Volume 24, Number 8—August 2018
Dispatch
Detection of Dengue Virus among Children with Suspected Malaria, Accra, Ghana
Figure 1
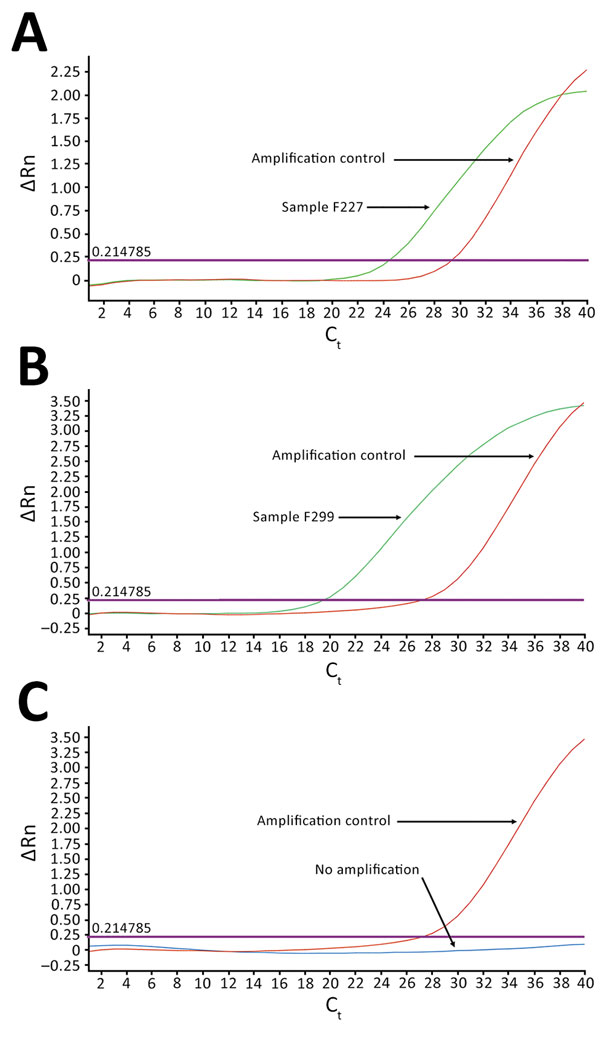
Figure 1. TaqMan array card amplification plots for 2 dengue virus–positive samples and 1 negative sample in study of dengue virus among 166 children with suspected malaria, Accra, Ghana, October 2016–July 2017. Blood samples (2.5 mL of the 5.0 mL collected) obtained from children reporting to the hospital with acute febrile illness (AFI) were screened for 26 pathogens simultaneously by using the real-time PCR TaqMan array card. The cards were in 384-well format, and each well contained 1 µL of reaction mixture (0.75 µL of the extracted total nucleic acid and 0.25 µL of TaqMan Fast Virus 1-step Master Mix; Life Technologies/Applied Biosystems, Foster City, CA, USA). The assays were run on an Applied Biosystems Quant Studio 7 Flex real-time PCR system, according to the manufacturer’s recommendations. Two samples tested positive for dengue virus: A) F227, which amplified with critical Ct of 24.40; and B) F299, which amplified with Ct of 19.35. C) All others tested negative, indicated by amplification signals (ΔRn) below threshold levels at quantification cycle cutoff of 35. Each assay included nucleic acid to serve as amplification controls. External controls were spiked into each to monitor extraction and amplification efficiency, and 1 negative control was included for each batch of extraction to monitor laboratory contamination. Ct, cycle threshold.