Volume 25, Number 11—November 2019
Research
Rare Detection of Bordetella pertussis Pertactin-Deficient Strains in Argentina
Cite This Article
Citation for Media
Abstract
Pertussis resurgence had been attributed to waning vaccine immunity and Bordetella pertussis adaptation to escape vaccine-induced immunity. Circulating bacteria differ genotypically from strains used in production of pertussis vaccine. Pertactin-deficient strains are highly prevalent in countries that use acellular vaccine (aP), suggesting strong aP-imposed selection of circulating bacteria. To corroborate this hypothesis, systematic studies on pertactin prevalence of infection in countries using whole-cell vaccine are needed. We provide pertussis epidemiologic data and molecular characterization of B. pertussis isolates from Buenos Aires, Argentina, during 2000–2017. This area used primary vaccination with whole-cell vaccine. Since 2002, pertussis case incidences increased at regular 4-year outbreaks; most cases were in infants <1 year of age. Of the B. pertussis isolates analyzed, 90.6% (317/350) contained the ptxP3-ptxA1-prn2-fim3-2 allelic profile. Immunoblotting and sequencing techniques detected only the 2 pertactin-deficient isolates. The low prevalence of pertactin-deficient strains in Argentina suggests that loss of pertactin gene expression might be driven by aP vaccine.
Vaccination against pertussis is mandatory worldwide. Two types of vaccines are currently in use: whole-cell vaccine (wP), which was the first vaccine developed, and acellular vaccine (aP), which contains purified components of Bordetella pertussis and was formulated because of adverse reactions associated with wP (1). Many countries continue to use wP for the primary vaccination series and for boosters recommended for children <7 years of age. Industrialized countries have switched to vaccination with aP. However, in the past 2 decades, the number of pertussis cases detected increased to ≈24.1 million/year; ≈161,000 deaths occurred during 2014 (2,3). Although most cases occur in developing countries, industrialized countries have also had large-scale outbreaks, even nations with high vaccination rates (2,4–6).
The main causes proposed for this changing pertussis epidemiology are vaccination coverage rates lower than the 90% recommended by the World Health Organization, waning of vaccine-induced immunity (7,8) (which occurs faster in the aP-vaccinated population), and evolution of circulating bacteria to vaccine immunity–evasive phenotypes (9,10). The first reports on bacterial evolution documented genetic polymorphisms encoding proteins included in the vaccines (e.g., pertactin [PRN], pertussis toxin, and the pertussis toxin promoter [ptxP]) (11,12). More recently, a major increase in the isolation of B. pertussis bacteria that do not express certain vaccine antigens was reported (10,13,14). In countries using PRN-containing aP vaccines, such as the United States, Canada, and Australia, the PRN-deficient isolates have increased substantially in the past 4 years (10,15,16). The expansion of strains deficient in PRN in populations vaccinated with PRN-containing aP vaccines indicates that such strains apparently have a selective advantage in populations vaccinated with aP vaccine (17).
To corroborate this hypothesis, we conducted systematic studies on PRN prevalence in Argentina, a country that uses wP vaccine. We monitored and analyzed B. pertussis population dynamics in Buenos Aires. Our aim was to assess whether PRN-deficient strains were circulating in Buenos Aires and to analyze the results obtained in relation to the vaccine used and the epidemiologic situation of the disease during 2000–2017.
Population Studied, Clinical Case Definition, and Laboratory Diagnosis
We used pertussis epidemiologic data and samples collected during 2000–2017 from the Pertussis Reference Laboratory (La Plata, Argentina). We collected data on patient sex, age, duration of symptoms, vaccination status, and laboratory results.
We confirmed clinical cases of pertussis in patients by isolation of B. pertussis culture, amplification of B. pertussis–specific DNA by using PCR, or serologic analysis (IgG titer to pertussis toxin >120 IU/mL). We defined a confirmed case of pertussis as one that meets the clinical case definition and is epidemiologically linked to a laboratory-confirmed case (18–20).
Vaccine Schedule in Buenos Aires
The wP vaccine was introduced in Argentina (population 44.9 million) during the 1970s and is still used for the 3 primary doses at 2, 4, and 6 months of age; for the 2 boosters at 18 months of age; and at school entry (5–6 years of age) in the public sector (≈90% of the population). The aP vaccine is used in the private sector and for the recommended boosters in adolescents, healthcare workers in contact with infants <12 months of age, household contacts of low-birthweight infants, and pregnant women.
In most of Argentina, the diphtheria–tetanus–pertussis vaccine is used as a third dose; coverage during recent years ranged from 91.0% to 95.0%; in certain jurisdictions, this value was <80.0% (21). Official coverage rates for adolescent boosters were 75.3% during 2015, 81.9% during 2016, and 88.0% during 2017. Official coverage rates for maternal immunization were 61.7% during 2015, 65.6% during 2016, and 67.0% during 2017.
Samples and Bacterial Growth Conditions
The Pertussis Reference Laboratory samples included nasopharyngeal specimens from 16,151 hospitalized patients from Buenos Aires with signs/symptoms of pertussis infection. These samples were routinely screened for B. pertussis by culture and PCR. B. pertussis culture was performed on Regan-Lowe agar (Difco, https://www.fishersci.com) supplemented with 15% (vol/vol) defibrinated fresh sheep blood at 36.5°C and monitored for 10 days. Suspected colonies were replicated in Bordet-Gengou agar (Difco) supplemented with 15% (vol/vol) defibrinated fresh sheep blood. Colonies exhibiting hemolysis were gram-stained and tested by using agglutination with B. pertussis–specific antiserum (Murex Diagnostic Products, https:///www.murex-ph.com) and PCR (22,23). The isolates were also biochemically typed by using the API-20-NE system (bioMérieux, https://www.biomerieux.com).
Isolates were stored at –80°C in 1% (wt/vol) casaminoacid solution containing 20% (vol/vol) glycerol. B. pertussis strain Tohama phase I (Collection de l’Institut Pasteur) was also grown on Bordet-Gengou agar at 36.5°C for 72 h.
B. pertussis Isolate Characterization
Genotyping
A total of 350 B. pertussis isolates collected in Buenos Aires during January 2000–December 2017 were included in the analyses (Table 1). For genetic typing, we amplified ptxP, pertussis toxin A subunit (ptxA), prn, and fimbriae type 3 (fim3) loci by using PCR with specific primers (Table 2) and sequenced them as described (24–32). We also screened isolates for an array of mutations causing deficiency in the immunogen PRN by using PCR amplification and molecular sequencing (26,27). We used primers 5′-CCCATTCTTCCCTGTTCCAT-3′ and 5′-CCTGAGCCTGGAGACTGG-3′ (27) to amplify the complete prn gene (27), and these primers in combination with internal primers to sequence the complete gene.
We calculated by year the discriminatory power of the multilocus sequence typing technique by using the equation reported by Hunter and Gaston (33). This equation is based on the probability that 2 unrelated strains sampled from the test population will be placed into different typing groups. Thus, the index can have any value between 0 and 1, with 0 representing the lowest discriminatory capacity, indicating that all the strains are in a single genotyping group (low diversity), and 1 representing the highest discriminatory capacity, indicating high genotypic diversity among the isolates.
PRN Immunoblotting
For this assay, we treated 2 ×1010 CFUs of B. pertussis isolates with Laemmli sample buffer, and subjected extracts to electrophoresis on 12.5% (wt/vol) sodium dodecyl sulfate–polyacrylamide gels. After electrophoresis, we transferred the proteins to a polyvinylidene phosphate membrane (Immobilon P; Millipore, http://www.emdmillipore.com) and incubated with a 1:2,500 dilution of PRN-specific polyclonal immune serum. This serum was obtained from BALB/c mice immunized with purified B. pertussis 69-kDa PRN (National Institute for Biological Standards and Control [NIBSC] code no. 90/654 version 4). We used alkaline phosphatase–labeled sheep anti-mouse immunoglobulins for detecting immune complexes and nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate as phosphatase chromogenic substrates (Biodynamics SRL, https://www.biodynamics.com.ar). The B. pertussis Tohama strain served as a PRN-positive control.
Serotype Analysis
We performed serotype analysis by using an agglutination assay with monoclonal antibodies (mABs) against fimbriae type 2 (anti-Fim2 mAb; NIBSC code no. 04/154) and fimbriae type 3 (anti-Fim3 mAb, NIBSC code no. 04/156). These analyses followed European Union laboratory group recommendations (34).
Pertussis Epidemiology in Buenos Aires
During 2000–2017, the Pertussis Reference Laboratory received 75% of total clinical samples (nasopharyngeal swab samples) from pertussis-suspected case-patients identified in Buenos Aires and reported to the Ministry of Health. Of these 16,151 samples, 3,220 (19.9%) were from laboratory-confirmed cases. A total of 2,870 samples were positive by PCR for B. pertussis–specific genes, and 350 samples were positive by PCR and culture.
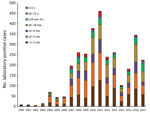
The provincial cases per year distribution reflected the pattern of the entire country; 3 outbreaks were detected, in 2008, 2011, and 2016 (Figure 1). In each year of the period analyzed, most cases were detected in infants <1–2 months of age and those >2–4 months of age (Figure 2). The high proportion of cases recorded in patients <6 months of age was expected because pertussis is most severe in that age group.
We obtained the distribution of confirmed pertussis cases by patient age and vaccination status. Of confirmed cases, data were complete for 72.6% (2,338/3,220) of vaccinated persons and for 26.5% (619/2,338) of nonvaccinated persons <2 months of age. For infants <6 months of age, 45.3% had complete age-specific vaccination schedules. The percentage of patients with complete schedules was 53.7% for children >6 months of age and 6.4% for adolescents >11 years of age. Although this percentage for adolescents was low, this age group contained considerably fewer persons than those <6 months of age (44 infants vs. 1,590 children >6 months of age).
B. pertussis Genotyping
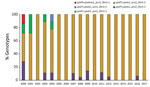
Almost all (99.7%) B. pertussis isolates analyzed contained the ptxA1 and prn2 alleles. Clinical isolates obtained during 2000–2004 had up to 4 different multilocus sequence typing genotypes (Figure 3). The index of discrimination calculated by year for this period ranged from 0.25 to 0.80. The highest value (higher diversity) was detected in 2000. The ptxP1 or ptxP4 variants were detected before 2004; thereafter, the ptxP3 locus prevailed. Most (291/350, 83.1%) isolates obtained after 2004 had the ptxP3-ptxA1-prn2-fim3–2 genotype. For 2004–2017, the index of discrimination ranged from 0 to 0.24, indicating the lowest diversity detected.
Fimbriae Serotyping
Of 350 isolates tested, only 1 obtained during 2016 was classified as Fim2. The remaining isolates were classified as Fim3.
PRN Immunoblots
Only 2 of the B. pertussis isolates included in this study were Prn deficient. Both strains were obtained from patients <1 year of age who had typical pertussis signs/symptoms. These 2 case-patients were linked in time (2016) but not geographically. One of these patients was born to a mother vaccinated with a PRN-containing aP vaccine and the other to a nonvaccinated mother. For these 2 strains, we detected insertion sequence 481 sequence (forward sense) at position 1613–1614 of prn, which disrupted the gene.
We conducted molecular genetic characterization of 350 B. pertussis isolates obtained during 2000–2017 from hospitalized patients in Buenos Aires, Argentina. Buenos Aires, similar to the entire country of Argentina, uses only wP vaccine for primary series of pertussis vaccinations. Most B. pertussis isolates were obtained during the outbreaks detected during 2007–2008 (83), 2011–2012 (145), and 2016–2017 (45). A total of 78% of isolates were obtained from patients <6 months of age, 13.7% from patients 6–12 months of age, and 8.3% from patients >12 months of age. As expected, most B. pertussis isolates were from unvaccinated persons. As detected in other countries, we found that almost all isolates characterized had the Fim3 serotype (35).
Of 350 isolates, variants ptxP1 and ptxP4 and the allele prn1 were detected before 2004. Starting in 2004, a total of 313 isolates obtained had the ptxP3-ptxA1-prn2 alleles with either fim3–1 or fim–3-2. These genotypes differed from those of the vaccine-production strains (36) and were the most common in other countries that had vaccinated populations (35).
The polymorphism in PRN and subsequent spread of PRN-deficient isolates have elicited a deep concern in the healthcare system because these changes hypothetically might represent a selective avoidance by the bacteria of the immunity induced by the vaccines. The predominance of prn2 detected in more recent isolates from Buenos Aires is consistent with the hypothesis that strains in the vaccinated population with that allele are fitter than those harboring other prn alleles (37).
Regarding a deficiency in PRN expression, we detected only 2 isolates containing insertion sequence 481 in the coding region of prn. These isolates were obtained from infants <1 year of age that were linked in time (2016) but not location. One of these patients was born to a mother vaccinated with a PRN-containing aP vaccine and the other patient was born to a nonvaccinated mother.
There are no reports of pertactin deficiency in a country such as Argentina that includes wP vaccine as the only vaccine for primary pertussis vaccination. Recently, Poland, the only country in Europe that still uses the wP vaccine but also the aP vaccine for primary series, has reported detection of PRN-deficient clinical isolates (38). The percentage of those isolates was lower (15%) than that detected in the United States, Canada, or Australia (>65%), which only use aP vaccines (10,39,40). Detection of these isolates might be a consequence of the increase in the use of aP vaccines in Poland. Within this context, our study is apparently unique because Argentina uses only wP vaccine for the primary series of pertussis vaccinations.
This low frequency of PRN-deficient strains in regions where wP vaccine is still used supports the hypothesis that PRN-deficient clinical isolates have an advantage in aP vaccine–primed immunity (41). Accordingly, PRN-deficient clinical isolates were able to overcome an anti-PRN–mediated inhibition of macrophage cytotoxicity in vitro (42). Moreover, this study also showed that recently collected PRN-deficient B. pertussis clinical isolates harboring a ptxP3 variant and the prn2 allele had higher CFUs per lung and were capable of sustaining infection longer in aP vaccine–immunized mice than isolates still producing the protein. The investigators of that study speculated that these particular isolates might be capable of infecting immunized persons at an earlier stage of waning immunity after aP vaccine immunization or postinfection, thus having an advantage over isolates producing PRN. The findings of Hegerle et al. (42) are consistent with those recently reported by Safarchi et al. (17), which indicated a higher fitness of Prn-negative strains in aP vaccine–immunized mice. These investigators demonstrated in a mixed-infection model that PRN-negative B. pertussis colonized the respiratory tract of aP-immunized mice more effectively than the PRN-positive strain, thus outcompeting that strain (17).
Regarding a possible association between clinical findings and the PRN expression of the bacterial isolates that caused the human infections, recent studies suggest that symptoms (with the exception of apneas, which were less likely in PRN-deficient infections) and clinical course were similar regardless of PRN expression (14,41). Clarke et al. provide new data on this subject, which suggest that rapid emergence of PRN-deficient B. pertussis variants is unlikely to contribute to any greater risk for death or severe outcomes from infections in young, vulnerable infants (43).
Our study supports the hypothesis regarding pathogen adaptation of B. pertussis to the type of vaccine used. A key finding in this study was that use of the wP vaccine in the primary series of vaccinations correlated with a near complete absence of PRN-deficient strains, although the aP vaccine was used in subsequent vaccine regimens. Continued surveillance for PRN production in circulating B. pertussis is needed, as well as monitoring of other possible genotypic changes in the B. pertussis population, including a lack of expression of other immunogens contained in acellular vaccines.
Mr. Carriquiriborde is a doctoral student at the Universidad Nacional de La Plata y Consejo Nacional de Investigaciones Científicas y Técnicas, La Plata, Argentina. His research interests are pertussis and vaccinology.
Acknowledgments
We thank Donald F. Haggerty for editing the final version of the manuscript.
This study was supported by the Agencia Nacional de Promoción Científica y Tecnológica, the Consejo Nacional de Investigaciones Científicas y Técnicas, and the Universidad Nacional de La Plata.
D.H. planned the study, interpreted data, and edited the figures and manuscript; D.B. interpreted data and edited the figures and manuscript; F.C., V.R., P.M.A., G.M., and E.B. performed experiments and laboratory analyses. All authors approved the final version of the manuscript.
References
- Yeung KHT, Duclos P, Nelson EAS, Hutubessy RCW. An update of the global burden of pertussis in children younger than 5 years: a modelling study. Lancet Infect Dis. 2017;17:974–80. DOIPubMedGoogle Scholar
- Clark TA. Changing pertussis epidemiology: everything old is new again. J Infect Dis. 2014;209:978–81. DOIPubMedGoogle Scholar
- Vizzotti C, Juarez MV, Bergel E, Romanin V, Califano G, Sagradini S, et al. Impact of a maternal immunization program against pertussis in a developing country. Vaccine. 2016;34:6223–8. DOIPubMedGoogle Scholar
- Falleiros Arlant LH, de Colsa A, Flores D, Brea J, Avila Aguero ML, Hozbor DF. Pertussis in Latin America: epidemiology and control strategies. Expert Rev Anti Infect Ther. 2014;12:1265–75. DOIPubMedGoogle Scholar
- Wendelboe AM, Van Rie A, Salmaso S, Englund JA. Duration of immunity against pertussis after natural infection or vaccination. Pediatr Infect Dis J. 2005;24(Suppl):S58–61. DOIPubMedGoogle Scholar
- McGirr A, Fisman DN. Duration of pertussis immunity after DTaP immunization: a meta-analysis. Pediatrics. 2015;135:331–43. DOIPubMedGoogle Scholar
- Mooi FR, Van Der Maas NA, De Melker HE. Pertussis resurgence: waning immunity and pathogen adaptation - two sides of the same coin. Epidemiol Infect. 2014;142:685–94. DOIPubMedGoogle Scholar
- Lam C, Octavia S, Ricafort L, Sintchenko V, Gilbert GL, Wood N, et al. Rapid increase in pertactin-deficient Bordetella pertussis isolates, Australia. Emerg Infect Dis. 2014;20:626–33. DOIPubMedGoogle Scholar
- Advani A, Gustafsson L, Ahrén C, Mooi FR, Hallander HO. Appearance of Fim3 and ptxP3-Bordetella pertussis strains, in two regions of Sweden with different vaccination programs. Vaccine. 2011;29:3438–42. DOIPubMedGoogle Scholar
- Kallonen T, Mertsola J, Mooi FR, He Q. Rapid detection of the recently emerged Bordetella pertussis strains with the ptxP3 pertussis toxin promoter allele by real-time PCR. Clin Microbiol Infect. 2012;18:E377–9. DOIPubMedGoogle Scholar
- Hegerle N, Guiso N. Bordetella pertussis and pertactin-deficient clinical isolates: lessons for pertussis vaccines. Expert Rev Vaccines. 2014;13:1135–46. DOIPubMedGoogle Scholar
- Bodilis H, Guiso N. Virulence of pertactin-negative Bordetella pertussis isolates from infants, France. Emerg Infect Dis. 2013;19:471–4. DOIPubMedGoogle Scholar
- Pawloski LC, Queenan AM, Cassiday PK, Lynch AS, Harrison MJ, Shang W, et al. Prevalence and molecular characterization of pertactin-deficient Bordetella pertussis in the United States. Clin Vaccine Immunol. 2014;21:119–25. DOIPubMedGoogle Scholar
- Tsang RS, Shuel M, Jamieson FB, Drews S, Hoang L, Horsman G, et al. Pertactin-negative Bordetella pertussis strains in Canada: characterization of a dozen isolates based on a survey of 224 samples collected in different parts of the country over the last 20 years. Int J Infect Dis. 2014;28:65–9. DOIPubMedGoogle Scholar
- Safarchi A, Octavia S, Luu LD, Tay CY, Sintchenko V, Wood N, et al. Pertactin negative Bordetella pertussis demonstrates higher fitness under vaccine selection pressure in a mixed infection model. Vaccine. 2015;33:6277–81. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Pertussis/whooping Cough (Bordetella pertussis). Case definition; 2014 [cited 2019 Aug 4]. https://wwwn.cdc.gov/nndss/conditions/pertussis/case-definition/2014
- World Health Organization. WHO-recommended surveillance standard of pertussis [cited 2019 Aug 4]. https://wwwwhoint/immunization/monitoring_surveillance/burden/vpd/surveillance_type/passive/pertussis_standards/en
- Argentina Ministry of Health. Whopping cough and convulsions of pertussis. Case definitions [in Spanish] [cited 2019 Aug 5]. http://www.msal.gob.ar/images/stories/pdf/coqueluche-recomendaciones-definiciones.pdf
- Gentile A. [Bordetella pertussis infection] [in Spanish]. Arch Argent Pediatr. 2010;108:78–81.PubMedGoogle Scholar
- Grimprel E, Bégué P, Anjak I, Betsou F, Guiso N. Comparison of polymerase chain reaction, culture, and western immunoblot serology for diagnosis of Bordetella pertussis infection. J Clin Microbiol. 1993;31:2745–50.PubMedGoogle Scholar
- Hozbor D, Fouque F, Guiso N. Detection of Bordetella bronchiseptica by the polymerase chain reaction. Res Microbiol. 1999;150:333–41. DOIPubMedGoogle Scholar
- Schouls LM, van der Heide HG, Vauterin L, Vauterin P, Mooi FR. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains reveals rapid genetic changes with clonal expansion during the late 1990s. J Bacteriol. 2004;186:5496–505. DOIPubMedGoogle Scholar
- Mooi FR, van Loo IH, van Gent M, He Q, Bart MJ, Heuvelman KJ, et al. Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis. 2009;15:1206–13. DOIPubMedGoogle Scholar
- Fiett J, Letowska I, Gniadkowski M, Hryniewicz W. The new strategy for allele identification of the genes coding for pertussis toxin subunit S1 (ptx S1) and pertactin (prn) in Bordetella pertussis. J Microbiol Methods. 2003;55:651–66. DOIPubMedGoogle Scholar
- Mooi FR, Hallander H, Wirsing von König CH, Hoet B, Guiso N. Epidemiological typing of Bordetella pertussis isolates: recommendations for a standard methodology. Eur J Clin Microbiol Infect Dis. 2000;19:174–81. DOIPubMedGoogle Scholar
- Borisova O, Kombarova SY, Zakharova NS, van Gent M, Aleshkin VA, Mazurova I, et al. Antigenic divergence between Bordetella pertussis clinical isolates from Moscow, Russia, and vaccine strains. Clin Vaccine Immunol. 2007;14:234–8. DOIPubMedGoogle Scholar
- Advani A, Donnelly D, Hallander H. Reference system for characterization of Bordetella pertussis pulsed-field gel electrophoresis profiles. J Clin Microbiol. 2004;42:2890–7. DOIPubMedGoogle Scholar
- Bottero D, Gaillard ME, Fingermann M, Weltman G, Fernández J, Sisti F, et al. Pulsed-field gel electrophoresis, pertactin, pertussis toxin S1 subunit polymorphisms, and surfaceome analysis of vaccine and clinical Bordetella pertussis strains. Clin Vaccine Immunol. 2007;14:1490–8. DOIPubMedGoogle Scholar
- van Loo IH, Heuvelman KJ, King AJ, Mooi FR. Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J Clin Microbiol. 2002;40:1994–2001. DOIPubMedGoogle Scholar
- Hardwick TH, Plikaytis B, Cassiday PK, Cage G, Peppler MS, Shea D, et al. Reproducibility of Bordetella pertussis genomic DNA fragments generated by XbaI restriction and resolved by pulsed-field gel electrophoresis. J Clin Microbiol. 2002;40:811–6. DOIPubMedGoogle Scholar
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–6.PubMedGoogle Scholar
- Network E-Ls. External quality assurance scheme for Bordetella identification and B. pertussis typing [cited 2019 Aug 4]. https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/EQA-pertussis-Bordetella-ID-B-typing.pdf
- Barkoff AM, Mertsola J, Pierard D, Dalby T, Hoegh SV, Guillot S, et al. Surveillance of circulating Bordetella pertussis strains in Europe during 1998 to 2015. J Clin Microbiol. 2018;56:e01998–17. DOIPubMedGoogle Scholar
- Bottero D, Gaillard ME, Basile LA, Fritz M, Hozbor DF. Genotypic and phenotypic characterization of Bordetella pertussis strains used in different vaccine formulations in Latin America. J Appl Microbiol. 2012;112:1266–76. DOIPubMedGoogle Scholar
- van Gent M, van Loo IH, Heuvelman KJ, de Neeling AJ, Teunis P, Mooi FR. Studies on Prn variation in the mouse model and comparison with epidemiological data. PLoS One. 2011;6:
e18014 . DOIPubMedGoogle Scholar - Polak M, Zasada AA, Mosiej E, Krysztopa-Grzybowska K, Witkowski L, Rzeczkowska M, et al. Pertactin-deficient Bordetella pertussis isolates in Poland: a country with whole-cell pertussis primary vaccination. Microbes Infect. 2018;Dec 21:pii: S11286-4579(18)30193-X.
- Quinlan T, Musser KA, Currenti SA, Zansky SM, Halse TA. Pertactin-negative variants of Bordetella pertussis in New York State: a retrospective analysis, 2004-2013. Mol Cell Probes. 2014;28:138–40. DOIPubMedGoogle Scholar
- Queenan AM, Cassiday PK, Evangelista A. Pertactin-negative variants of Bordetella pertussis in the United States. N Engl J Med. 2013;368:583–4. DOIPubMedGoogle Scholar
- Martin SW, Pawloski L, Williams M, Weening K, DeBolt C, Qin X, et al. Pertactin-negative Bordetella pertussis strains: evidence for a possible selective advantage. Clin Infect Dis. 2015;60:223–7. DOIPubMedGoogle Scholar
- Hegerle N, Dore G, Guiso N. Pertactin deficient Bordetella pertussis present a better fitness in mice immunized with an acellular pertussis vaccine. Vaccine. 2014;32:6597–600. DOIPubMedGoogle Scholar
- Clarke M, McIntyre PB, Blyth CC, Wood N, Octavia S, Sintchenko V, et al. The relationship between Bordetella pertussis genotype and clinical severity in Australian children with pertussis. J Infect. 2016;72:171–8. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: September 30, 2019
1These authors contributed equally to this article.
Table of Contents – Volume 25, Number 11—November 2019
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Daniela Hozbor, Laboratorio VacSal, Instituto de Biotecnología y Biología Molecular, Departamento de Ciencias Biológicas, Facultad de Ciencias Exactas, Universidad Nacional de La Plata y Centro Científico Tecnológico–La Plata, Consejo Nacional de Investigaciones Científicas y Técnicas, Calles 47 y 115, 1900 La Plata, Argentina; or
Top