Volume 25, Number 12—December 2019
Dispatch
Distantly Related Rotaviruses in Common Shrews, Germany, 2004–2014
Figure 1
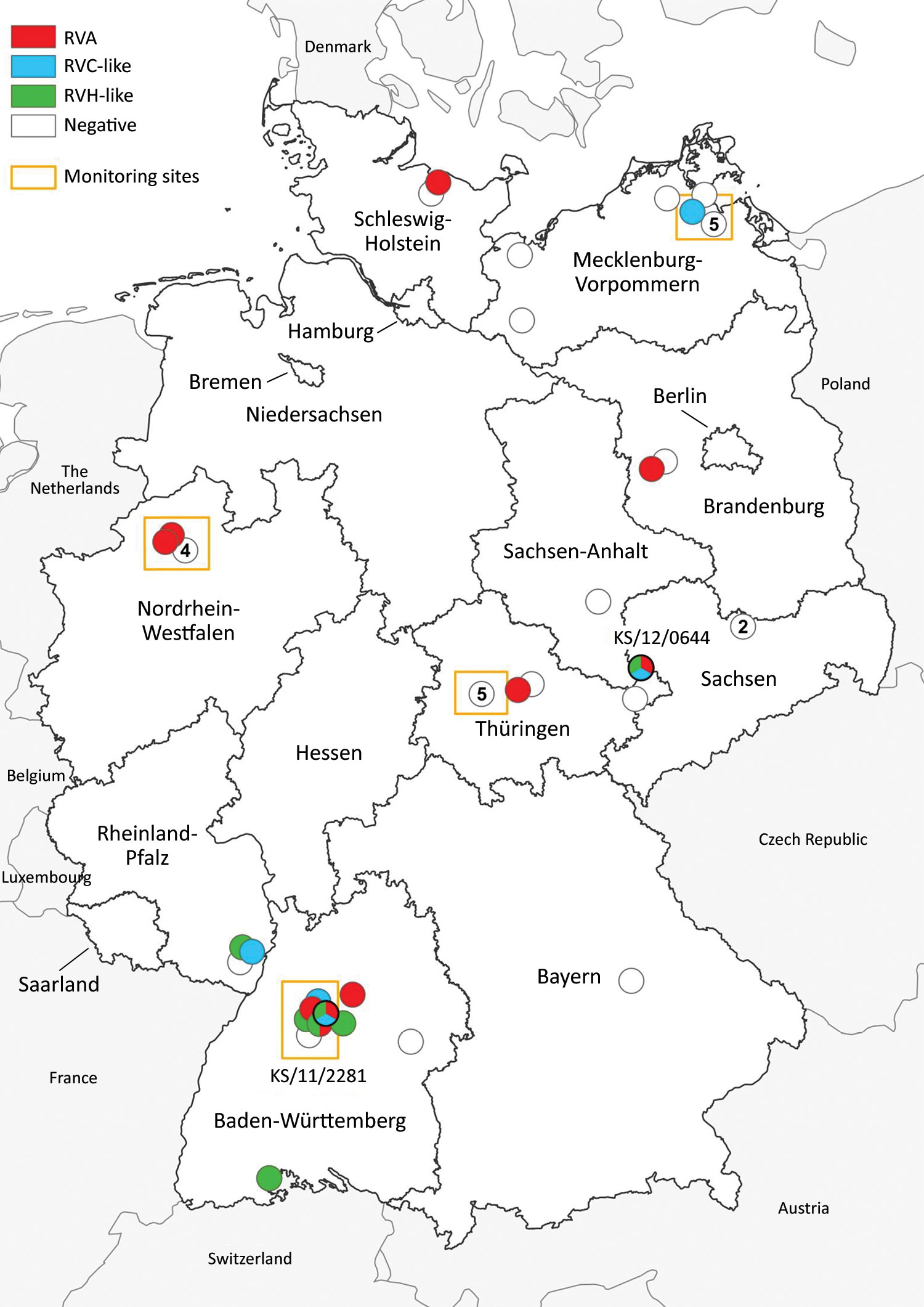
Figure 1. Distribution of common shrews (Sorex araneus) collected at monitoring sites (9) and additional sites (10) in Germany, 2004–2014, positive and negative for RVA, RVC-like, and RVH-like species by reverse transcription PCR. Numbers in white circles indicate the number of negative samples at that collection site; white circles without numbers indicate 1 negative sample at that site. Circles with multiple colors indicate animals with co-infections. The collection sites of the 2 samples analyzed in detail by next-generation sequencing (KS/12/0644 and KS/11/2281; tricolored circles) are indicated. RVA, rotavirus A; RVC, rotavirus C; RVH, rotavirus H.
References
- Tate JE, Burton AH, Boschi-Pinto C, Parashar UD; World Health Organization–Coordinated Global Rotavirus Surveillance Network. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin Infect Dis. 2016;62(Suppl 2):S96–105. DOIPubMedGoogle Scholar
- Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, et al. Family: Reoviridae. In: King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ, editors. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Amsterdam: Elsevier Academic Press; 2012. p. 541–637.
- Bányai K, Kemenesi G, Budinski I, Földes F, Zana B, Marton S, et al. Candidate new rotavirus species in Schreiber’s bats, Serbia. Infect Genet Evol. 2017;48:19–26. DOIPubMedGoogle Scholar
- International Committee on Taxonomy of Viruses. Taxonomic information. Virus taxonomy: 2018b release. 2018 Jul [cited 2019 Aug 21]. https://talk.ictvonline.org/taxonomy
- Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Van Ranst M, Johne R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Arch Virol. 2012;157:1177–82. DOIPubMedGoogle Scholar
- Matthijnssens J, Ciarlet M, Rahman M, Attoui H, Bányai K, Estes MK, et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch Virol. 2008;153:1621–9. DOIPubMedGoogle Scholar
- Martella V, Bányai K, Matthijnssens J, Buonavoglia C, Ciarlet M. Zoonotic aspects of rotaviruses. Vet Microbiol. 2010;140:246–55. DOIPubMedGoogle Scholar
- Schlegel M, Radosa L, Rosenfeld UM, Schmidt S, Triebenbacher C, Löhr PW, et al. Broad geographical distribution and high genetic diversity of shrew-borne Seewis hantavirus in Central Europe. Virus Genes. 2012;45:48–55. DOIPubMedGoogle Scholar
- Fischer S, Mayer-Scholl A, Imholt C, Spierling NG, Heuser E, Schmidt S, et al. Leptospira genomospecies and sequence type prevalence in small mammal populations in Germany. Vector Borne Zoonotic Dis. 2018;18:188–99. DOIPubMedGoogle Scholar
- Mayer-Scholl A, Hammerl JA, Schmidt S, Ulrich RG, Pfeffer M, Woll D, et al. Leptospira spp. in rodents and shrews in Germany. Int J Environ Res Public Health. 2014;11:7562–74. DOIPubMedGoogle Scholar
- Scheuch M, Höper D, Beer M. RIEMS: a software pipeline for sensitive and comprehensive taxonomic classification of reads from metagenomics datasets. BMC Bioinformatics. 2015;16:69. DOIPubMedGoogle Scholar
- Tausch SH, Renard BY, Nitsche A, Dabrowski PW. RAMBO-K: rapid and sensitive removal of background sequences from next generation sequencing data. PLoS One. 2015;10:
e0137896 . DOIPubMedGoogle Scholar - Wang XX, Li JX, Wang WG, Sun XM, He CY, Dai JJ. [Preliminary investigation of viruses to the wild tree shrews (Tupaia belangeri Chinese)]. Dongwuxue Yanjiu. 2011;32:66–9.PubMedGoogle Scholar
- Li K, Lin XD, Huang KY, Zhang B, Shi M, Guo WP, et al. Identification of novel and diverse rotaviruses in rodents and insectivores, and evidence of cross-species transmission into humans. Virology. 2016;494:168–77. DOIPubMedGoogle Scholar
- Otto PH, Rosenhain S, Elschner MC, Hotzel H, Machnowska P, Trojnar E, et al. Detection of rotavirus species A, B and C in domestic mammalian animals with diarrhoea and genotyping of bovine species A rotavirus strains. Vet Microbiol. 2015;179:168–76. DOIPubMedGoogle Scholar
Page created: November 18, 2019
Page updated: November 18, 2019
Page reviewed: November 18, 2019
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.