Volume 25, Number 6—June 2019
Dispatch
New Delhi Metallo-β-Lactamase 5–Producing Klebsiella pneumoniae Sequence Type 258, Southwest China, 2017
Figure 2
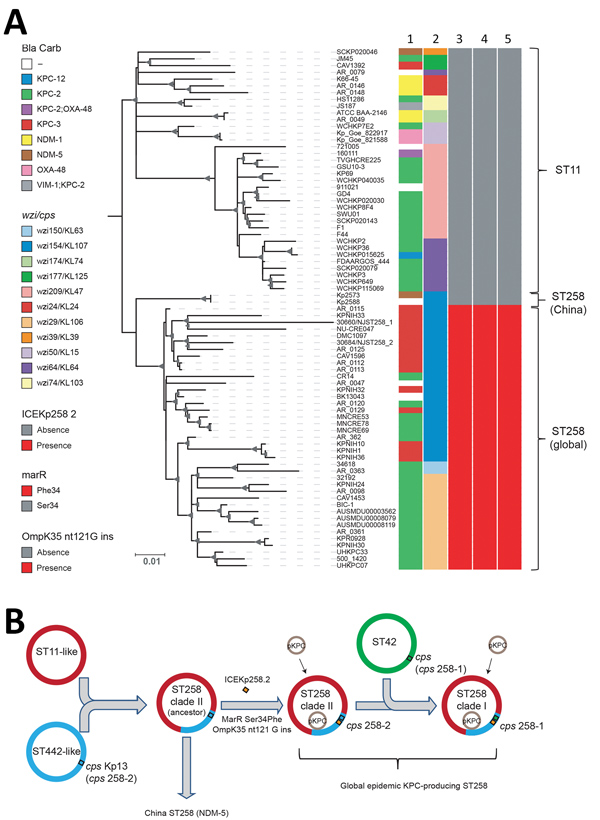
Figure 2. Phylogenetic analysis of KPC-producing and NDM-5–producing CG258 Klebsiella pneumoniae strains from China, 2017, and reference strains. A) Core SNP phylogenetic analysis of 76 global CG258 (ST258 and ST11) and 2 ST258 strains from China. Lane 1, Bla_carb; lane 2, wzi (cps); lane 3, integrative and conjugative element Kp258.2; lane 4, marR; lane 5, ompK35 gene (guanine insertion at nt position 121). The maximum-likelihood tree was rooted by using ST11 strains. Bootstrap values >90% are indicated as gray triangles at branch points. Sizes are proportion to values. Scale bar indicates nucleotide substitutions per site. B) Updated hypothesis of the molecular evolution of carbapenem-resistant K. pneumoniae ST258 (9). Bla, β-lactamase; Carb, carbapenemase; cps, capsular polysaccharide gene; ICE, integrative and conjugative element; ins, insertion; KPC, Klebsiella pneumoniae carbapenemase; MarR, transcriptional regulator protein of the multiple antimicrobial drug resistance repressor family; NDM, New Delhi metallo-β-lactamase; ompK35, outer membrane protein K35; OXA, oxacillinase; pKPC, plasmid carrying KPC; ST, sequence type; VIM-1, Verona integron–encoded metallo- β-lactamase 1; wzi, surface assembly of capsule gene.
References
- Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant cones. Trends Microbiol. 2016;24:944–56. DOIPubMedGoogle Scholar
- Patel G, Bonomo RA. “Stormy waters ahead”: global emergence of carbapenemases. Front Microbiol. 2013;4:48. DOIPubMedGoogle Scholar
- Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–96. DOIPubMedGoogle Scholar
- Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. DOIPubMedGoogle Scholar
- Lascols C, Peirano G, Hackel M, Laupland KB, Pitout JD. Surveillance and molecular epidemiology of Klebsiella pneumoniae isolates that produce carbapenemases: first report of OXA-48-like enzymes in North America. Antimicrob Agents Chemother. 2013;57:130–6. DOIPubMedGoogle Scholar
- Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41:252–75. DOIPubMedGoogle Scholar
- Mathers AJ, Peirano G, Pitout JD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28:565–91. DOIPubMedGoogle Scholar
- Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59:5873–84. DOIPubMedGoogle Scholar
- Chen L, Mathema B, Pitout JD, DeLeo FR, Kreiswirth BN. Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio. 2014;5:e01355–14. DOIPubMedGoogle Scholar
- Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, et al. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS One. 2015;10:
e0133727 . DOIPubMedGoogle Scholar
1These authors contributed equally to this article.