Volume 25, Number 9—September 2019
Dispatch
Use of Human Intestinal Enteroids to Detect Human Norovirus Infectivity
Abstract
Tools to detect human norovirus infectivity have been lacking. Using human intestinal enteroid cultures inoculated with GII.Pe-GII.4 Sydney–infected fecal samples, we determined that a real-time reverse transcription PCR cycle threshold cutoff of 30 may indicate infectious norovirus. This finding could be used to help guide infection control.
Human norovirus accounts for 18% of acute gastroenteritis cases worldwide (1). Molecular nucleic acid tests, such as real-time reverse transcription PCR (rRT-PCR), are widely used for laboratory diagnosis of norovirus RNA in clinical samples (2). These molecular assays are virus specific and their analytical sensitivity is high, but they cannot distinguish between infectious and noninfectious viruses. To study the correlation between viral load measured by rRT-PCR and virus infectivity, we used a recently developed human intestinal enteroid (HIE) culture system (cultures that contain multiple intestinal epithelial cell types) for human norovirus. Ethics approval for this study was obtained from the Joint Chinese University of Hong Kong–New Territories East Cluster Clinical Research Ethics Committee (reference no. 2016.516).
We examined the infectivity of human pandemic norovirus genogroup II genotype 4 (GII.Pe-GII.4 Sydney) strains at different inoculating levels by using the adult stem cell–derived HIE line J2 as described previously (3). We seeded enteroid monolayers on 96-well cell culture plates at a density of 6–8 × 104 cells/well and maintained them in differentiation media for 5 days before being inoculated with norovirus. We used fecal samples from 3 norovirus-positive children and from adults in our norovirus surveillance program in Hong Kong (4). We prepared 1% fecal suspensions and filtered them once by using 0.22-μm centrifugal filters, then prepared 3-fold serial dilutions of fecal filtrates in infection medium and stored them at −70°C in multiple aliquots for single use. These dilutions mimicked a broad range of cycle threshold (Ct) values of a widely used diagnostic rRT-PCR to represent high to very low norovirus levels (5). We used 33 μL of each dilution of fecal filtrate (brought to 100 μL in infection medium) for inoculation of each dilution and performed all virus inoculation steps and downstream cell cultures on enteroids with <20 passages in the presence of bile acid (glycochenodeoxycholic acid). We measured norovirus RNA levels in supernatant at 1, 24, and 72 h after inoculation by using rRT-PCR with a 10-fold serially diluted standard of in vitro–transcribed norovirus RNA. We considered a >10-fold increase in RNA level at 72 h after inoculation from baseline (1 h after inoculation) to indicate productive viral replication and to confirm the presence of infectious virus. We also subjected fecal filtrate dilutions to norovirus antigen detection by use of the Food and Drug Administration–cleared commercial RIDASCREEN Norovirus 3rd Generation EIA (R-Biopharm AG, https://clinical.r-biopharm.com), according to the manufacturer’s instructions. We used the same amount of fecal filtrate for inoculation into HIE and EIA measurement and compared the analytical sensitivity of HIE and EIA.
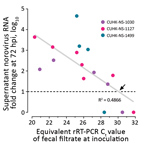
We selected 3 strains of norovirus GII.Pe-GII.4 Sydney, and all replicated productively in HIE line J2; the maximum fold increase of norovirus RNA ranged from 120 (2.1 log10) to 45,793 (4.7 log10). Higher levels of replication were obtained from fecal samples from adults (Figure 1). We identified no virus replication when cells were inoculated with fecal filtrate dilutions with Ct >30. The Ct values of the inoculating virus dilution that exhibited a transition from a positive-to-negative enteroid culture result (i.e., having a 10-fold RNA increase) were 27.7, 29.0, and 30.0 for each of the 3 strains. By using linear regression analysis on pooled data of the 3 strains, we estimated that a Ct cutoff of <30.1 in inoculating fecal filtrates would indicate the ability to generate productive norovirus GII.Pe-GII.4 Sydney replication (i.e., containing infectious norovirus) (Figure 2).
From 2014 through 2018, a total of 114 (6.5%) of 1,754 norovirus-positive fecal samples from patients admitted to the Prince of Wales Hospital, Sha Tin, Hong Kong, with acute gastroenteritis had Ct values >30 (Ct median 17.8; interquartile range 14.8–22.3; range 5.5–39.2). Among the 1,579 (90.0%) genotyped samples, the proportion of GII.4 was 49.1%; other GII, 44.8%; GI, 5.2%; and co-infections with >1 norovirus capsid genotype, 0.9%. Analytical sensitivity of virus replication in HIEs for measuring moderate norovirus shedding was higher than that of EIA by being able to detect infectious virus in fecal filtrate dilutions with Ct values of 25–30 (Table).
The limitations of highly sensitive molecular nucleic acid tests and the clinical value of proving virus infectivity were demonstrated in a recent study of Zika virus infection that found that the virus could be detected by culturing samples with high viral RNA levels only (6). For human norovirus, laboratory tools to detect its infectivity have been lacking. Recent technologic advancement in culturing human norovirus in HIE provides a chance to examine norovirus infectivity in laboratory settings (3). Using pandemic norovirus GII.Pe-GII.4 Sydney strains, we showed that the most sensitive method for detecting human norovirus was rRT-PCR, followed by HIE infection; EIA was the least sensitive. We experimentally determined that norovirus inocula with Ct <30 can robustly yield productive virus replication in HIE, suggesting the presence of infectious virus. This value concurs with findings of a study that proposed an optimal Ct cutoff of 31 when attributing a disease to norovirus by comparing between symptomatic and asymptomatic cases (7). Our findings imply that a small proportion (≈6.5%) of patients shedding low levels of norovirus RNA (Ct >30) may not be infectious.
In an experimental human volunteer challenge study with norovirus GII.4, low levels of viral RNA shedding were found 10 days after challenge (8). HIE infection can now be used to reevaluate archived samples to better define parameters that would correlate with the infectious period of norovirus gastroenteritis to guide infection control. Of note, in a meta-analysis, the prevalence of norovirus RNA shedding from asymptomatic patients was estimated to be ≈9% (9). Moreover, in a large-scale study of outbreak data from CaliciNet (https://www.cdc.gov/norovirus/reporting/calicinet/index.html), a national norovirus surveillance network in the United States, the median Ct value among asymptomatic patients was 28 (10). We hypothesize that a substantial proportion of asymptomatic patients are shedding infectious norovirus and that their role in spreading the virus merits our attention. We have shown that high levels of norovirus replication can be achieved from fecal samples of adults, not just young children, in HIE cultures (11).
The use of a fixed Ct cutoff in clinical context needs to be interpreted with caution. First, neither a standardized protocol to perform rRT-PCRs nor a World Health Organization International Standard for norovirus is available to harmonize assay variability across laboratories worldwide. Differences in recovery among nucleic acid extraction methods may further complicate reproducible determination of Ct values. Second, virus culture systems in cell lines generally lack sensitivity (12), and that of HIE remains unknown. However, it is probably not optimal because input genome equivalents for norovirus to achieve replication are not extremely low (50% infectious dose 4.4 × 102 to 2.1 × 103 copies/well) (3,11). It is possible that samples with Ct >30 might still contain infectious virus and that low amounts of replicating norovirus would only be detected with further serial propagation of the virus. Third, norovirus replication varies between samples and virus genotypes in HIE culture (11).
In summary, we demonstrate that a Ct cutoff of 30 for a widely used clinical diagnostic rRT-PCR can indicate the presence of infectious GII.Pe-GII.4 Sydney norovirus in an HIE culture model. Patients shedding low levels of norovirus RNA may not be infectious, which should be considered both for estimation of attributable norovirus burden and for clinical management of viral gastroenteritis.
Dr. Chan is an assistant professor in the Department of Microbiology and Stanley Ho Centre for Emerging Infectious Diseases and a principal investigator in the Li Ka Shing Institute of Health Sciences of the Chinese University of Hong Kong. His research interest focuses on molecular epidemiology and pathogenesis of human noroviruses; other research interests include use of metagenomic next-generation sequencing in clinical diagnostics, gut virome, and food virology.
Acknowledgments
This work was supported in part by the Hong Kong Research Grants Council (to M.C.-W.C.; reference no. 14162217), a seed fund for gut microbiota research by the Faculty of Medicine of the Chinese University of Hong Kong (to P.K.S.C.), and a Public Health Service grant (to M.K.E.; reference no. PO1 AI057788).
M.K.E. is named as an inventor on patents related to cloning of the Norwalk virus genome and is a consultant to and has received support from Takeda Vaccines, Inc. Other authors have no conflict of interests to declare.
References
- Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis. 2014;14:725–30. DOIPubMedGoogle Scholar
- Vinjé J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol. 2015;53:373–81. DOIPubMedGoogle Scholar
- Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–93. DOIPubMedGoogle Scholar
- Chan MC, Kwok K, Zhang LY, Mohammad KN, Lee N, Lui GCY, et al. Bimodal seasonality and alternating predominance of norovirus GII.4 and non-GII.4, Hong Kong, China, 2014–2017. Emerg Infect Dis. 2018;24:767–9. DOIGoogle Scholar
- Kageyama T, Kojima S, Shinohara M, Uchida K, Fukushi S, Hoshino FB, et al. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41:1548–57. DOIPubMedGoogle Scholar
- Mead PS, Duggal NK, Hook SA, Delorey M, Fischer M, Olzenak McGuire D, et al. Zika virus shedding in semen of symptomatic infected men. N Engl J Med. 2018;378:1377–85. DOIPubMedGoogle Scholar
- Phillips G, Lopman B, Tam CC, Iturriza-Gomara M, Brown D, Gray J. Diagnosing norovirus-associated infectious intestinal disease using viral load. BMC Infect Dis. 2009;9:63. DOIPubMedGoogle Scholar
- Bernstein DI, Atmar RL, Lyon GM, Treanor JJ, Chen WH, Jiang X, et al. Norovirus vaccine against experimental human GII.4 virus illness: a challenge study in healthy adults. J Infect Dis. 2015;211:870–8. DOIPubMedGoogle Scholar
- Qi R, Huang YT, Liu JW, Sun Y, Sun XF, Han HJ, et al. Global prevalence of asymptomatic norovirus infection: a meta-analysis. EClinicalMedicine. 2018;2-3:50–8.
- Shioda K, Barclay L, Becker-Dreps S, Bucardo-Rivera F, Cooper PJ, Payne DC, et al. Can use of viral load improve norovirus clinical diagnosis and disease attribution? Open Forum Infect Dis. 2017;4:ofx131.
- Costantini V, Morantz EK, Browne H, Ettayebi K, Zeng XL, Atmar RL, et al. Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg Infect Dis. 2018;24:1453–64. DOIPubMedGoogle Scholar
- Leland DS, Ginocchio CC. Role of cell culture for virus detection in the age of technology. Clin Microbiol Rev. 2007;20:49–78. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: July 30, 2019
Table of Contents – Volume 25, Number 9—September 2019
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Martin C.-W. Chan, Prince of Wales Hospital, Department of Microbiology, 1/F, Lui Che Woo Clinical Sciences Bldg, Shatin, Hong Kong, China
Top