Volume 26, Number 11—November 2020
CME ACTIVITY - Research
Phage-Mediated Immune Evasion and Transmission of Livestock-Associated Methicillin-Resistant Staphylococcus aureus in Humans
Figure 1
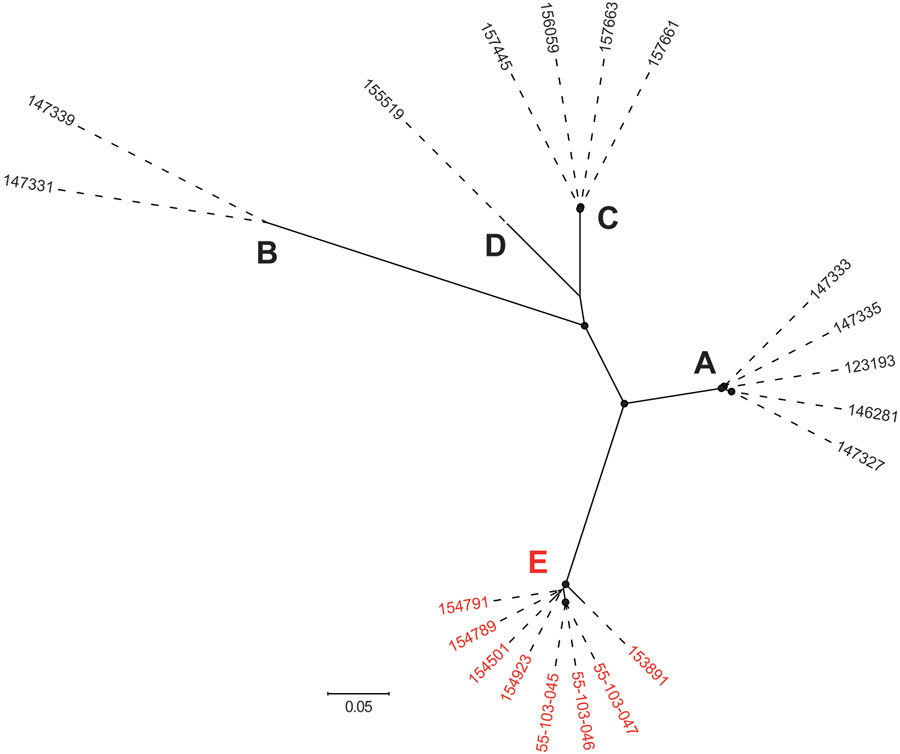
Figure 1. Maximum-likelihood phylogeny showing the genetic diversity of the 20 IEC-harboring ΦSa3int prophages identified in livestock-associated methicillin-resistant Staphylococcus aureus CC398 isolates from North Denmark Region, Denmark. Capital letters indicate phylogenetic clusters (A–E). Red text indicates ΦSa3int prophages harboring both immune evasion cluster and tarP (cluster E). The phylogeny was estimated for 795 high-quality core SNPs. Filled circles at nodes indicate bootstrap values >90%. Scale bar represents number of nucleotide substitutions per variable site.
1These senior authors contributed equally to this article.
Page created: September 11, 2020
Page updated: October 15, 2020
Page reviewed: October 15, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.