Volume 26, Number 11—November 2020
Dispatch
Multiple Introductions of Salmonella enterica Serovar Typhi H58 with Reduced Fluoroquinolone Susceptibility into Chile
Figure 2
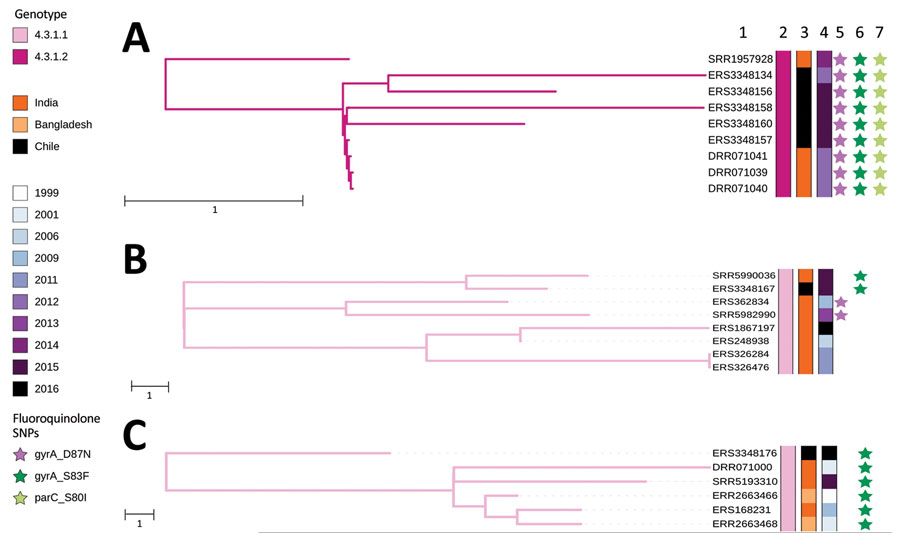
Figure 2. Nearest-neighbor calculations of Salmonella enterica serovar Typhi of genotype 4.3.1 and maximum-likelihood phylogenetic trees for 3 introductions of Salmonella Typhi genotype 4.3.1 into Chile in the context of their closest Salmonella Typhi isolate neighbors. A) Isolate collected during 2012–2014 resembles isolates from South Asia. B) Isolate collected during 2015 resembles isolates from India. C) Isolate collected in 2016 is closely related to a cluster of sequences from India and Bangladesh. Accession numbers, genotypes, countries, and years of isolation are shown. Stars indicate mutations in the quinolone resistance determining region of genes gyrA, gyrA-S83F and gyrA-D87N, and parC-S80I. Scale bars indicate SNP distance. SNP, single-nucleotide polymorphism.
1These first authors contributed equally to this work.
2These authors contributed equally to this work.