Volume 26, Number 11—November 2020
Dispatch
Multiple Introductions of Salmonella enterica Serovar Typhi H58 with Reduced Fluoroquinolone Susceptibility into Chile
Cite This Article
Citation for Media
Abstract
Salmonella enterica serovar Typhi H58, an antimicrobial-resistant lineage, is globally disseminated but has not been reported in Latin America. Genomic analysis revealed 3 independent introductions of Salmonella Typhi H58 with reduced fluoroquinolone susceptibility into Chile. Our findings highlight the utility of enhanced genomic surveillance for typhoid fever in this region.
Salmonella enterica serovars Typhi, Paratyphi A, and Paratyphi B are the etiologic agents of typhoid and paratyphoid fever. Each year, ≈11–21 million cases and 128,000–161,000 typhoid-related deaths occur, making typhoid a continued health concern in many low- and middle-income countries, particularly among populations without access to clean water or improved sanitation (1). Salmonella Typhi H58 lineage, genotype 4.3.1, commonly is associated with multidrug resistance, including resistance to chloramphenicol, ampicillin, and trimethoprim/sulfamethoxazole. In addition, isolates exhibiting resistance to fluoroquinolones have been linked to emergent clades of genotype 4.3.1 in South Asia (2), the spread of which could cause major challenges for disease management.
Salmonella Typhi H58 4.3.1 is the dominant genotype in many parts of Southeast and South Asia and in East Africa (3) and has spread globally but has not been reported in Latin America. Recent data on typhoid fever in South America are limited, and little is known about the population structure and antimicrobial susceptibility profiles of Salmonella Typhi on the continent. However, a report of 402 Salmonella Typhi isolates collected in Colombia during 2012–2015 showed that only 2.2% were resistant to fluoroquinolones (4). In 2016, Colombia reported collecting 204 Salmonella Typhi isolates, 12.7% of which exhibited decreased susceptibility to fluoroquinolones (5). Because these reports did not include whole-genome sequencing (WGS) data, determining whether isolates were genotype 4.3.1 is not possible.
Before the 1970s, typhoid fever was endemic in parts of South America and hyperendemic in Chile. However, water quality and sanitation improvements across the continent, partly in response to a major cholera epidemic in 1991, likely have contributed to a steep decline in the incidence of typhoid fever (6). During 1982–1992, Chile implemented interventions to reduce typhoid fever, including immunizing schoolchildren, prohibiting use of untreated sewage to irrigate crops, and detecting and treating chronic carriers. These interventions drastically reduced transmission and typhoid incidence has declined to 0.2 cases/100,000 persons (7), including in the greater Santiago metropolitan region (8).
Chile’s epidemiologic surveillance system tracks suspected typhoid fever. Two thirds of cases are confirmed by pathogen isolation from ordinarily sterile body fluids, such as blood or bone marrow. Salmonella Typhi isolates from Chile typically are susceptible to antimicrobial agents, but ciprofloxacin resistance has been reported. Among isolates collected during 2009–2016, nearly 2% were ciprofloxacin resistant and 14% displayed intermediate resistance (9). We used WGS and bioinformatic analyses to characterize Salmonella Typhi isolates from Chile to determine if antimicrobial-resistant H58 4.3.1 isolates have been introduced into South America.
We used a HiSeq WGS platform (Illumina, https://www.illumina.com) to generate 150 bp paired-end reads from Salmonella Typhi isolates collected during 2011–2017 by Chile’s National Typhoid Surveillance System. We assigned sequences to previously defined genotypes and identified 7 genotype 4.3.1 isolates (Appendix 1). Isolates were obtained from clinical cases in the Santiago metropolitan region: 1 in 2012, 5 in 2015, and 1 in 2016. For global context, we analyzed these 7 genomes and 2,386 publicly available sequences (Appendix 2 Table 1). Among publicly available sequences, 2,326 were genotype 4.3.1 and 60 were non–4.3.1 genotypes (Appendix 1 Table 2). We used the non–4.3.1 genotypes and a Salmonella Paratyphi A sequence as an outgroup for phylogenetic tree rooting. We produced clean and filtered SNP alignments (Appendix 1) and used these alignments to infer maximum likelihood phylogenies and specified a generalized time-reversible model and a Gamma distribution to model site-specific rate variation by using GTRGAMMA in RAxML version 8.2.9 (https://github.com/stamatak/standard-RAxML) and 100 bootstrap pseudoreplicates to assess branch support. SNP distances were calculated by using snp-dists (https://github.com/tseemann/snp-dists; Appendix 2). Raw Illumina reads were assembled by using either Velvet version 1.2 (European Bioinformatics Institute, https://www.ebi.ac.uk/~zerbino/velvet) or Unicycler version 0.4.7 (10). Assembled reads were input into Pathogenwatch (https://pathogen.watch) to detect nonsynonymous mutations in the quinolone-resistance determining region of gyrA and parC genes responsible for reduced fluoroquinolone susceptibility. We also used this approach to look for known antimicrobial resistance (AMR) genes. We further screened the sequences from Chile and close genetic relatives to determine molecular determinants of AMR and known plasmid replicon genes (Appendix 1 Table).
Phylogenomic and SNP analyses confirmed 7 Salmonella Typhi genotype 4.3.1 isolates from Chile. Contact tracing implies that 4/5 isolates from 2015 were part of a localized outbreak. We found that the 7 isolates were members of 2 different sublineages, lineage I (4.3.1.1) and lineage II (4.3.1.2), suggesting multiple introductions into Chile. The 2 isolates of lineage I carried a single gyrA-S83F mutation predicted to confer reduced susceptibility to fluoroquinolones. The 5 lineage II isolates carried 3 quinolone-resistance determining region mutations, 2 in gyrA genes, S83F and D87N, and 1 in parC-S80I. Genotype 4.3.1 triple mutants were predicted to be resistant to fluoroquinolones, and isolates of this sublineage with identical mutations have been observed on the subcontinent of India and have been associated with treatment failure (2,11,12). None of the lineage II triple mutants in Chile carried detectable horizontally acquired AMR genes.
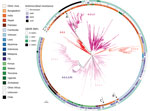
To provide a global contextualization of Salmonella Typhi genotype 4.3.1 in Chile, we analyzed the novel sequences alongside 2,326 existing sequences from 31 countries (Figure 1). The 4.3.1.2 triple mutants from Chile formed a closely related phylogenetic cluster (median distance 2 SNPs) with sequences that have the same antimicrobial susceptibility profile isolated from India during 2012–2014, indicating an introduction from South Asia (Figure 2, panel A).
The two 4.3.1.1 isolates from 2015 and 2016 in Chile were in distinct subclades of the tree and were separated by 19 SNPs, suggestive of 2 separate introductions. Of these, 1 introduction was closely related to a 2015 isolate from India (5 SNPs apart) (Figure 2, panel B) and the other was nested in a cluster of sequences from Southeast and South Asia and most closely related (median distance of 20 SNPs) to sequences from India and Bangladesh (Figure 2, panel C).
Our study confirmed Salmonella Typhi H58 genotype 4.3.1 in South America. Phylogenomic and SNP analyses indicate >3 separate genotype introductions into Chile; 5/7 isolates carried 3 distinct mutations, 2 in the gyrA gene, at D87N and S83F, and 1 in the parC gene at S80I, which are associated with ciprofloxacin resistance. For a high-income country with adequate surveillance, like Chile, the presence of fluoroquinolone-resistant genotype 4.3.1 Salmonella Typhi has no immediate implications. However, if this genotype is transferred to low- or middle-income countries in South America, it could have major consequences. Therefore, these data should be of concern to other countries in the region where potential typhoid fever transmission remains high and adequate sanitation might be lacking (5,6,10). Ciprofloxacin is a first-line drug for typhoid fever in much of Latin America, and fluoroquinolone-resistant genotype 4.3.1 would reduce its long-term efficacy.
Most diagnostic laboratories across South America are using pulsed-field gel electrophoresis to study Salmonella Typhi epidemiology (13), but efforts are underway to implement WGS for epidemiologic surveillance in several countries (14,15). However, WGS-based approaches for detecting genotype 4.3.1 and understanding trends in genotype population, circulating lineages, and AMR dynamics have not been adopted widely across the region. Our work highlights the need for a uniform WGS platform for global Salmonella Typhi monitoring and the need to elucidate the current epidemiology of typhoid fever in South America.
Ms. Maes is a PhD student at the University of Cambridge. Her research interests include the intracellular pathogen Salmonella Typhi, phylogenetics of Salmonella Typhi in Latin America, and the behavior of Salmonella Typhi in macrophages and the gallbladder.
Acknowledgments
The authors thank Jennifer Jones for her assistance with isolation of genomic DNA, the Wellcome Sanger Institute for facilitating sequencing, and Nick Thompson for enabling access to the Sanger cluster and pipelines.
This work was supported by The Wellcome Trust (STRATAA grant nos. 106158 and 098051) and the Gates Foundation (TyVAC grant no. OPP1151153). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
M.M. and G.D. receive funding from the National Institute for Health Research (NIHR) of the Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health and Social Care.
References
- World Health Organization. Typhoid [cited 2020 Apr 16]. https://www.who.int/health-topics/typhoid
- Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, Feasey NA, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet. 2015;47:632–9. DOIPubMedGoogle Scholar
- Park SE, Pham DT, Boinett C, Wong VK, Pak GD, Panzner U, et al. The phylogeography and incidence of multi-drug resistant typhoid fever in sub-Saharan Africa. Nat Commun. 2018;9:5094. DOIPubMedGoogle Scholar
- Diaz-Guevara P, Montaño LA, Duarte C, Zabaleta G, Maes M, Martinez Angarita JC, et al. Surveillance of Salmonella enterica serovar Typhi in Colombia, 2012-2015. PLoS Negl Trop Dis. 2020;14:
e0008040 . DOIPubMedGoogle Scholar - Pan American Health Organization. Epidemiological alert, Salmonella enterica serovar Typhi haplotype H58, 18 October 2018 [cited 2020 Apr 16]. https://iris.paho.org/handle/10665.2/50533
- Marco C, Delgado I, Vargas C, Muñoz X, Bhutta ZA, Ferreccio C. Typhoid fever in Chile 1969–2012: Analysis of an epidemic and its control. Am J Trop Med Hyg. 2018;99(3_Suppl):26–33.
- Government of Chile, Department of Epidemiology. Quarterly epidemiological bulletin 2015 Jan 13 [in Spanish] [cited 2020 Apr 16]. http://epi.minsal.cl/wp-content/uploads/2016/01/FT_BET2_2015.pdf
- Gauld JS, Hu H, Klein DJ, Levine MM. Typhoid fever in Santiago, Chile: Insights from a mathematical model utilizing venerable archived data from a successful disease control program. PLoS Negl Trop Dis. 2018;12:
e0006759 . DOIPubMedGoogle Scholar - de Salud M. Chile; Instituto de Salud Pública. Antimicrobial resistance bulletin 2018 [in Spanish] [cited 2020 Apr 16]. http://www.ispch.cl/sites/default/files/BoletinRam2018-08012019A.pdf
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Wong VK, Baker S, Connor TR, Pickard D, Page AJ, Dave J, et al.; International Typhoid Consortium. An extended genotyping framework for Salmonella enterica serovar Typhi, the cause of human typhoid. Nat Commun. 2016;7:12827. DOIPubMedGoogle Scholar
- Pham Thanh D, Karkey A, Dongol S, Ho Thi N, Thompson CN, Rabaa MA, et al. A novel ciprofloxacin-resistant subclade of H58 Salmonella Typhi is associated with fluoroquinolone treatment failure. eLife. 2016;5:
e14003 . DOIPubMedGoogle Scholar - Campos J, Pichel M, Vaz TMI, Tavechio AT, Fernandes SA, Muñoz N, et al. Building PulseNet Latin America and Caribbean Salmonella regional database: first conclusions of genetic subtypes of S. Typhi, S. Typhimurium and S. Enteritidis circulating in six countries of the region. Food Res Int. 2012;45:1030–6. DOIGoogle Scholar
- Baker KS, Campos J, Pichel M, Della Gaspera A, Duarte-Martínez F, Campos-Chacón E, et al. Whole genome sequencing of Shigella sonnei through PulseNet Latin America and Caribbean: advancing global surveillance of foodborne illnesses. Clin Microbiol Infect. 2017;23:845–53. DOIPubMedGoogle Scholar
- Chinen I, Campos J, Dorji T, Pérez Gutiérrez E. PulseNet Latin America and the Caribbean network: present and future. Foodborne Pathog Dis. 2019;16:489–97. DOIPubMedGoogle Scholar
Figures
Cite This Article1These first authors contributed equally to this work.
2These authors contributed equally to this work.
Table of Contents – Volume 26, Number 11—November 2020
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Mailis Maes, Cambridge Institute of Therapeutic Immunology & Infectious Disease Medicine, University of Cambridge, Puddicombe Way, Cambridge, Cambridgeshire CB2 0AW, UK
Top