Volume 26, Number 12—December 2020
Dispatch
Outbreaks of Highly Pathogenic Avian Influenza (H5N6) Virus Subclade 2.3.4.4h in Swans, Xinjiang, Western China, 2020
Figure 1
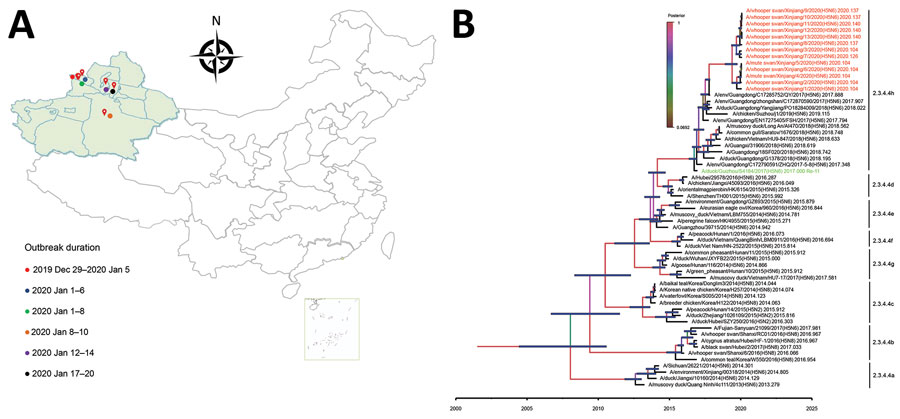
Figure 1. Geography and phylogeny of avian influenza (H5N6) outbreaks among migratory whooper swans (Cygnus cygnus) and mute swans (C. olor), Xinjiang Province, China, January 2020. A) Disease outbreak sites are marked with red drops, and dates of the outbreaks are indicated. Inset map shows islands in the South China Sea. B) Phylogenetic tree of the hemagglutinin (HA) genes of H5 viruses. The HA gene maximum clade credibility tree of the H5 viruses was constructed by using the BEAST 1.8.4 software package (https://beast-dev.github.io/beast-mcmc). Node bars indicate 95% highest posterior density of the node height. Each branch is colored by posterior probability: the 13 H5N6 viruses reported in this study are shown in red and the HA donor of the H5N1 vaccine Re-11 in green. The time to the most recent common ancestor is labeled at the bottom of the tree, which was estimated by using the Bayesian Markov chain Monte Carlo method in the BEAST 1.8.4 software package.