Volume 26, Number 2—February 2020
Research
Unique Clindamycin-Resistant Clostridioides difficile Strain Related to Fluoroquinolone-Resistant Epidemic BI/RT027 Strain
Figure 1
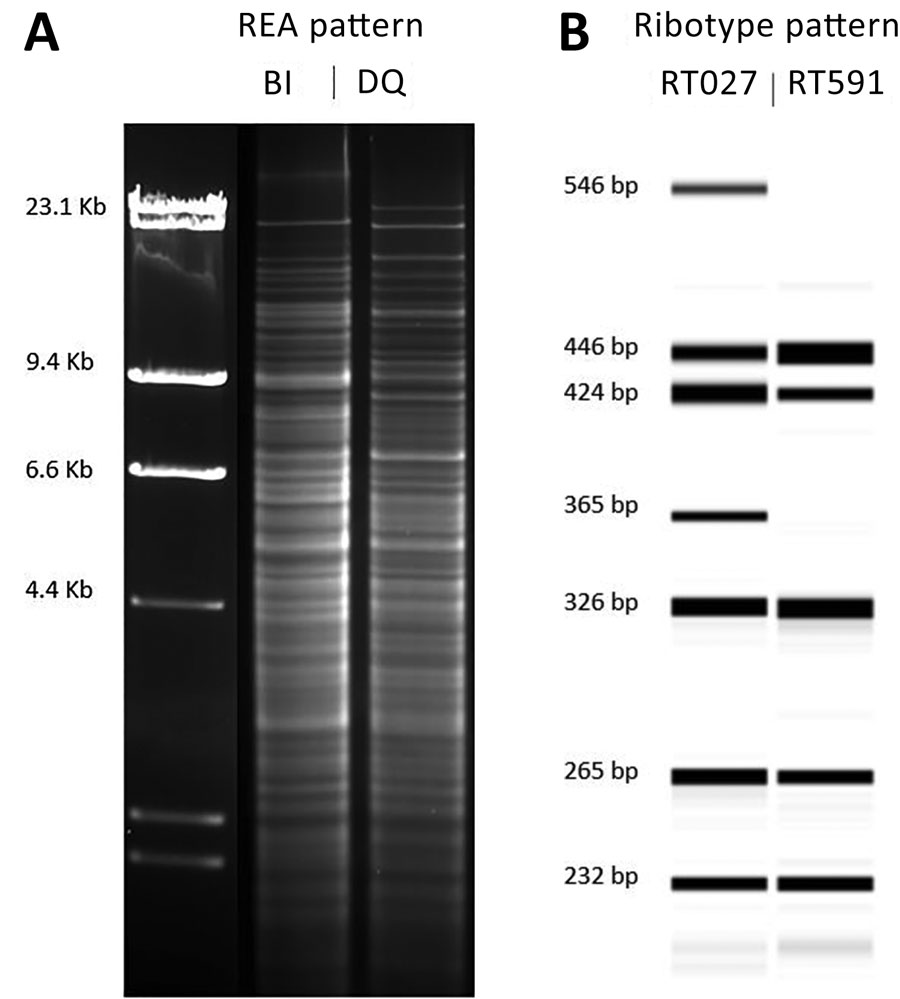
Figure 1. Comparison of the molecular characteristics of Clostridioides difficile strain DQ/591 and the epidemic BI/027 strain in study of C. difficile at 2 US Veteran Affairs long-term care facilities and their affiliated acute care facilities. The HindIII REA (A) and PCR ribotype (B) banding patterns were distinct between REA strain DQ/RT591 and REA strain BI/RT027. Molecular weight markers (in kb) are shown adjacent to the REA gel pattern. An internal spiked LIZ 1200 standard was used for fragment length calibrations (in bp) of the PCR ribotype gel patterns. REA, restriction endonuclease analysis.
Page created: January 17, 2020
Page updated: January 17, 2020
Page reviewed: January 17, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.