Volume 26, Number 4—April 2020
Dispatch
Person-to-Person Transmission of Andes Virus in Hantavirus Pulmonary Syndrome, Argentina, 2014
Figure 2
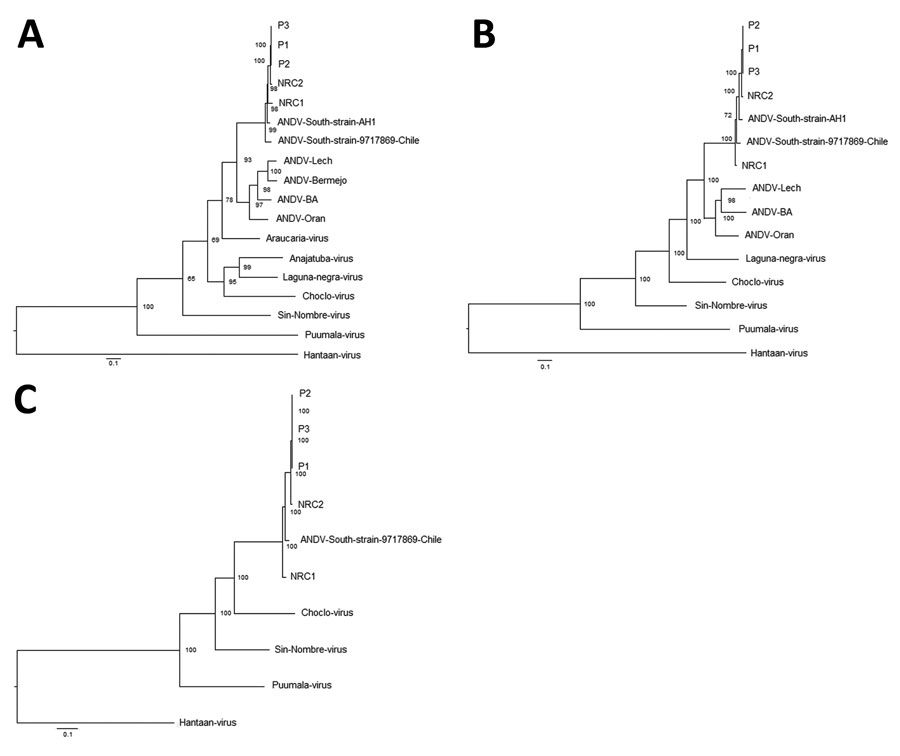
Figure 2. Phylogenetic analysis of hantaviruses based on complete genome of Andes virus (ANDV) isolated from case-patients in Argentina, 2014, and other orthohantaviruses characterized previously. A) Small (S) segment; B) medium (M) segment; C) large (L) segment. We used MrBayes version 3.2.7 (https://nbisweden.github.io/MrBayes) to reconstruct Bayesian maximum clade credibility trees. Numbers along branches are bootstrap values. Bootstrap support was based on 1,000 maximum-likelihood replicates. Scale bars indicate nucleotide substitutions per site. GenBank accession nos.: ANDV-South P1, S: MN850083, M: MN850088, L: MN850093; ANDV-South P2, S: MN850084, M: MN850089, L: MN850094; ANDV-South P3, S: MN850085, M: MN850090, L: MN850095; ANDV-South NRC1, S: MN850086, M: MN850091, L: MN850096; ANDV-South NRC2, S: MN850087, M: MN850092, L: MN850097; ANDV-Orán, S: AF325966, M: AF028024; Laguna Negra virus, S: NC038505, M: NC038506; ANDV-South, S: AF004660, M: AF324901; ANDV-South strain 9717869 Chile, S: AF291702, M: AF291703, L: AF291704; ANDV-Lech, S: AF482714, M: AF028022; ANDV-Bermejo, S: AF482713; Araucaria virus, S: AY740633; Anajatuba virus, S: JX443690; Choclo virus, S: KT983771, M: KT983772, L: EF397003; Sin Nombre virus, S: NC_005216, M: NC_005215, L: NC_005217; Puumala virus, S: NC_005224, M: NC_005223, L: NC_005225; Hantaan virus, S: JQ083395, M: JQ083394, L: JQ083393.