Volume 26, Number 4—April 2020
Research
Isolation of Drug-Resistant Gallibacterium anatis from Calves with Unresponsive Bronchopneumonia, Belgium
Figure 2
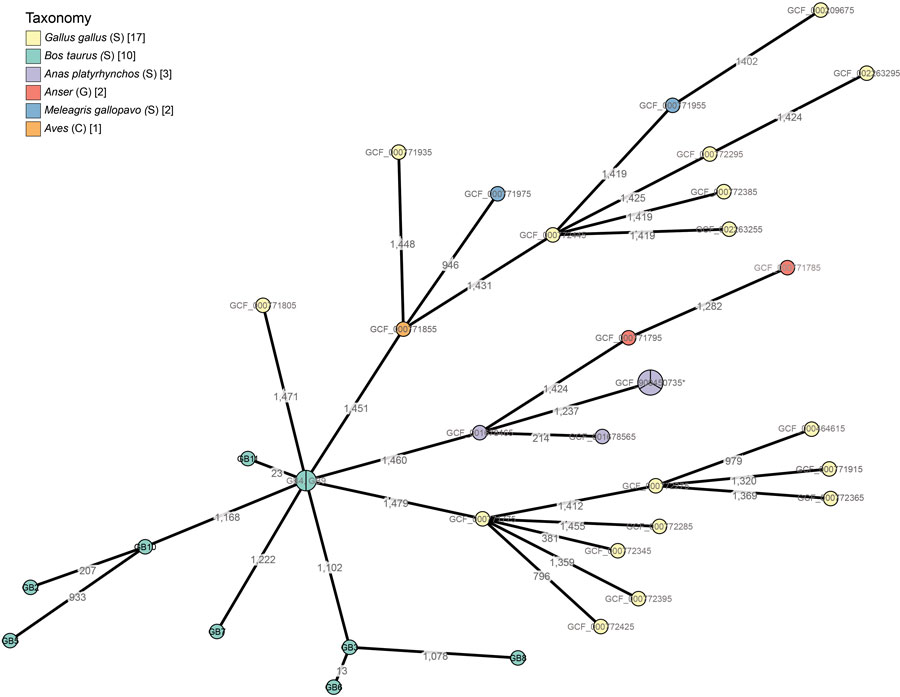
Figure 2. Phylogeny of Gallibacterium anatis isolates from cattle in Belgium, 2017–2018, based on a core genome multilocus sequence typing scheme constructed by using the 10 cattle isolates and 27 poultry isolates from National Center for Biotechnology Information (1,516 loci in total). Branch lengths are scaled logarithmically, and branch labels express number of allelic differences between isolates. Nodes scale with the number of isolates that have the same core genome multilocus sequence type. Nodes are colored according to the host organism of the isolate. Asterisk indicates node containing samples GCF_000379785, GCF_000772265, and GCF_900450735 (GB3, GB6, GB8) with the same sequence type. C; class; G, genus; S, species.
Page created: March 17, 2020
Page updated: March 17, 2020
Page reviewed: March 17, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.