Volume 26, Number 6—June 2020
Research
Antimicrobial Resistance in Salmonella enterica Serovar Paratyphi B Variant Java in Poultry from Europe and Latin America
Figure 2
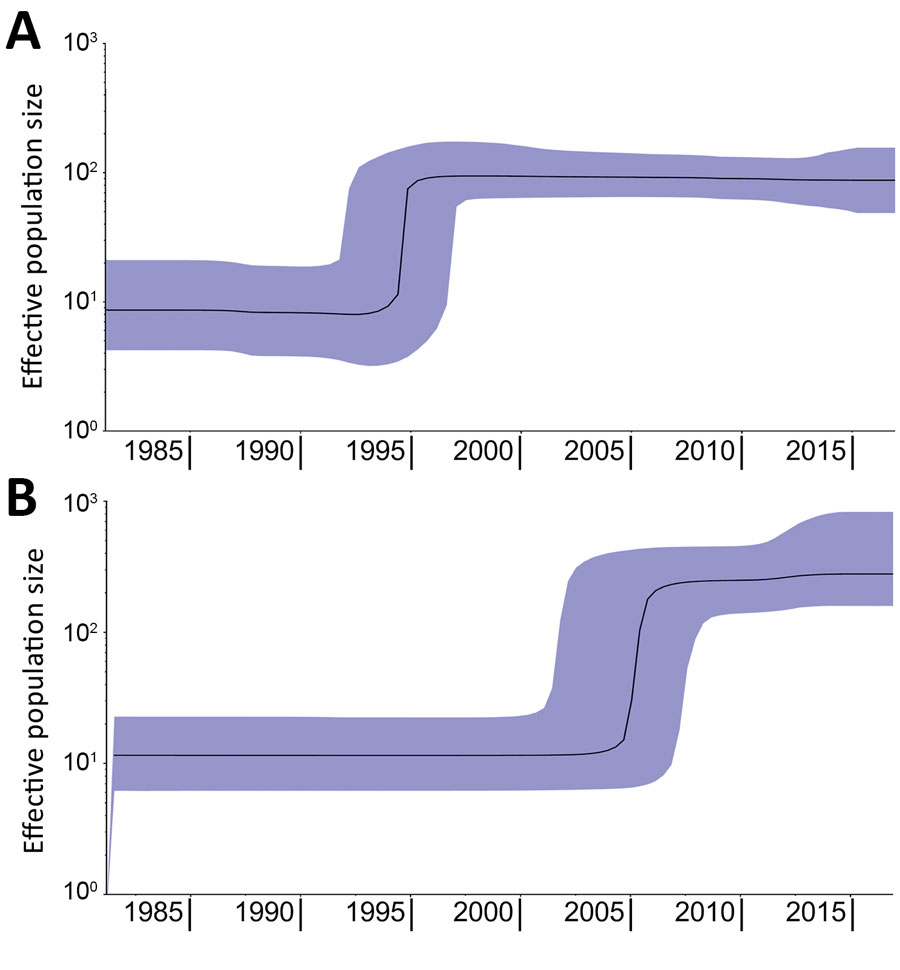
Figure 2. Bayesian skyline plots showing increase in effective population size of Salmonella enterica serovar Paratyphi B variant Java sequence type 28. Plots were made separately with strains originating from Europe (A) or Latin America (B). Emergence in Europe occurred in ≈1995 and in Latin America in ≈2005. Black lines indicate estimates of the median population over time; purple shading indicates 95% CIs.
1Current affiliate: Quadram Institute Bioscience, Norwich, UK.
2These authors contributed equally to this article.
Page created: May 18, 2020
Page updated: May 18, 2020
Page reviewed: May 18, 2020
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.