Volume 26, Number 6—June 2020
Research
Emergence of New Non–Clonal Group 258 High-Risk Clones among Klebsiella pneumoniae Carbapenemase–Producing K. pneumoniae Isolates, France
Cite This Article
Citation for Media
Abstract
The worldwide spread of Klebsiella pneumoniae carbapenemase–producing Klebsiella pneumoniae (KPC-Kp) isolates was reported to be caused by dissemination of 1 clonal complex (i.e., clonal group [CG] 258, which includes sequence types [STs] 258 and 512). We conducted whole-genome sequencing and epidemiologic analysis of all KPC-Kp isolates in France in 2018 and found that new successful high-risk clones of ST147, ST307, ST231, and ST383 are now the main drivers of blaKPC genes. The blaKPC genes were mostly carried by Tn4401a and Tn4401d structures and a new non–Tn4401 element. Our epidemiologic investigations showed that the emergence of these non-CG258 KPC-Kp isolates in France was linked to dissemination of these clones from Portugal. Thus, KPC-Kp epidemiology has changed in Europe, at least in several non–KPC-endemic countries of western Europe, such as France and Portugal, where CG258 is not the most prevalent clone.
In Klebsiella pneumoniae bacteria, resistance to carbapenems results in 2 main mechanisms: the production of an extended spectrum β-lactamase or plasmid-borne cephalosporinase associated with a decrease in permeability of the outer membrane (especially through alteration of OmpK35 and OmpK36 porins), or the production of a carbapenemase (1,2). In France, these carbapenemases are Ambler’s class A KPC enzymes; class B metallo-β-lactamases of NDM-, VIM- and, to a lesser extent, IMP-type; and Ambler’s class D oxacillinases of OXA-48–like type (3,4).
K. pneumoniae carbapenemase (KPC) was first identified in United States in the early 2000s (5). Since then, this carbapenemase has spread and has become endemic in several countries, including the United States, Israel, Greece, China, and Italy. It has also been sporadically described in many countries of Europe (1). The worldwide spread of KPC has been linked to the dissemination of a main clone of K. pneumoniae (sequence type [ST] 258) and a single-locus variant (ST512) (6). In Asia (especially China), ST11, another single-locus variant of ST258, is mostly reported among blaKPC-harboring K. pneumoniae isolates (7). In addition, a recently published study, conducted by the EUSCAPE working group in 2013 in Europe, revealed that the spread of carbapenemase-producing K. pneumoniae was driven by only a few clones (8). The most prevalent carbapenemase was KPC (45.5% [311/684 isolates]), and 72.7% (229/311) of KPC-producing K. pneumoniae (KPC-Kp) belong to the same clonal group (CG) 258, including ST258 and ST512. Whole-genome sequencing (WGS) analysis has suggested that ST258 and ST512 KPC-Kp spread out in Europe from 2 KPC-endemic countries: Greece (ST258) and Italy (ST512) (6,9–11). However, that study described the epidemiology of KPC in Europe in 2013, whereas the aim of our study was to describe the genomic characteristics of KPC isolates from a more recent period.
Analysis of the genetic context of blaKPC has revealed that this gene is mostly localized into a class 2 transposon named Tn4401 (12). Several variants of this Tn4401 (Tn4401a through Tn4401i) have been reported with deletions upstream of blaKPC within the promoter region (13,14). Consequently, expression of blaKPC genes is complex and might involve different promoters, depending on the specific genetic environment and bacterial species. The 2 main promoters are named P1, which is in the vicinity of blaKPC, and P2, a hybrid promoter located partly in the inverted repeat right of ISKpn7 (15). In rare cases, blaKPC genes have been described in genetic structure not related to Tn4401 and are named non–Tn4401 elements (NTE) (16). However, in NTE, the expression of blaKPC is mediated by other promoters. Our study aimed to deeply characterize the epidemiology of KPC-Kp circulating in France in 2018.
Strains Collections and Culture Conditions
We included all KPC-Kp sent to France’s National Reference Center for Antimicrobial Resistance during January 1–December 31, 2018. As previously described, we used isolates that were recovered from clinical and screening specimens and sent on a voluntary basis by any type of laboratory related to any health facility, such as private and public hospitals, nursing homes, and community laboratories (3,4). These laboratories were located throughout France, including overseas territories. KPC-Kp recovered by the National Reference Center for Antimicrobial Resistance represent ≈85%–90% of the KPC-Kp infection cases reported to the French Public Health Agency (R.A. Bonnin, L. Dortet, unpub. data). The collection used for WGS analysis represents a total of 63 nonduplicate isolates recovered from rectal screening (n = 45), urine (n = 12), blood cultures (n = 1), wound infections (n = 2), and respiratory samples (n = 2) and 1 isolate for which no recovery site information was available. Because the aim of the study was to evaluate the genetic diversity of KPC producers, we discarded from further analysis any duplicate isolates or isolates recovered from the same patient.
Antimicrobial Susceptibility Testing and Carbapenemase Detection
We performed antimicrobial susceptibility testing by using the disc diffusion method on Mueller-Hinton agar (Bio-Rad, https://www.bio-rad.com) and interpreted results according to European Committee on Antimicrobial Susceptibility Testing guidelines as updated in 2018 (http://www.eucast.org). We determined MICs for colistin by using broth microdilution (Sensititer Thermofisher, https://www.thermofisher.com). We performed carbapenemase detection by using Rapidec Carba NP (bioMérieux, https://www.biomerieux.com), followed by immunochromatographic detection of the carbapenemase enzyme using NG-Carba5 test (NG Biotech, https://ngbiotech.com).
WGS and Bioinformatic analysis
We sequenced all KPC-Kp isolates by using Illumina technology as previously described (17). We extracted total DNA from colonies by using the Ultraclean Microbial DNA Isolation Kit (MO BIO Laboratories, https://www.mobio.com) according to the manufacturer’s instructions. We prepared the DNA library as previously described (17) and performed de novo assembly and read mappings by using CLC Genomics Workbench 12.0 (QIAGEN, https://www.qiagen.com). We identified the acquired antimicrobial resistance genes by using Resfinder 3.1 (https://cge.cbs.dtu.dk/services/ResFinder/) and the CARD database (https://card.mcmaster.ca). We annotated the genomes by using RAST (18). We performed phylogenic analysis by using CSIphylogeny 1.4 (https://cge.cbs.dtu.dk/services/CSIPhylogeny) and visualized the genomes by using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree). We performed sequences alignments by using ClustalW (https://www.genome.jp/tools-bin/clustalw). We analyzed single-nucleotide polymorphisms (SNPs) on the whole genome by using CSIphylogeny V1.4 with parameters as follows: select minimum depth at SNP position at 10×, minimum distance between SNPs at 10 bp, and minimum SNP quality score of 30.
We constructed the genetic contexts by using de novo assembly or by mapping with reference genomes from GenBank and verified by in-house PCR as previously described (17). We analyzed plasmid contents of clinical isolates by using PlasmidFinder 2.1 to search for the replicase gene and by conducting manual searches for genes showing homology with the replicase gene.
Low Prevalence of KPC Producers among Carbapenem-Resistant K. pneumoniae in France
In 2018, a total of 3,931 carbapenem-resistant Enterobacteriaceae were collected, including 1,259 carbapenem-resistant K. pneumoniae, among which 1,010 were carbapenemase producers. OXA-48-like enzymes were the most prevalent carbapenemases (69.4%), followed by NDM (17.1%); 37 isolates (3.7%) had a combination of both of these carbapenemases. KPC enzymes represent only 3.0% of all carbapenemases, corresponding to 6.8% (69 isolates, including 6 duplicated isolates) of all carbapenemase produced by K. pneumoniae. KPC also was produced by 13 non–K. pneunomiae, including 5 KPC-2 producers (2 Escherichia coli, 1 Klebsiella oxytoca, 1 Enterobacter cloacae, and 1 Citrobacter koseri) and 8 KPC-3 producers (5 E. coli, 1 C. freundii, 1 E. cloacae, and 1 K. aerogenes). Accordingly, K. pneumoniae is the most prevalent species (84.1%) among KPC producers.
Antimicrobial-Susceptibility of KPC-Kp
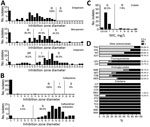
Susceptibility testing revealed that all KPC-Kp were resistant to all broad-spectrum cephalosporins (ceftazidime, cefotaxime, cefepime) and monobactam (Figure 1). All KPC-Kp were resistant to ertapenem. According to European Committee on Antimicrobial Susceptibility Testing breakpoints, 62.1% (41/66) KPC-Kp isolates remained susceptible to imipenem and 30.3% (20/66) to meropenem (Figure 1). Ceftazidime/avibactam (98.5% susceptibility) and colistin (92.2%) remained the most potent agents (Figure 1, panels B and C). However, we identified 1 isolate resistant to ceftazidime/avibactam but susceptible to carbapenems (Figure 1). This isolate produces a new variant of KPC, named KPC-39, that has been reported to possess increased ceftazidime catalytic activity but also to have concomitantly lost its carbapenemase activity (19). Among other antimicrobial families, 84.9% KPC-Kp isolates were susceptible to tigecycline, 30.3% to sulfamethoxazole/trimethoprim, and 19.7% to ciprofloxacin (Figure 1, panel D). Resistance to aminoglycosides varied from 40.9% for amikacin to 75.8% for tobramycin.
blaKPC Variants and Associated Acquired Resistance Genes
We performed WGS on 63 nonduplicate KPC-Kp isolates and identified their resistomes by using Illumina technology. In this collection, 44 isolates possessed the blaKPC-3 gene (69.8%), and 18 (28.5%) possessed the blaKPC-2 gene. One isolate harbored a novel single-nucleotide variant of blaKPC-3, blaKPC-39 (Figure 1, panel B). Two isolates produced 2 carbapenemases, including 1 isolate coharboring blaKPC-2 and blaVIM-1 and another 1 coharboring blaKPC-2 and blaNDM-4 (Figure 2). We identified additional antimicrobial-resistance determinants in all isolates (Appendix 1). Approximately 46% of the KPC-Kp isolates carried an extended spectrum β-lactamase encoding gene, including 22 isolates harboring blaCTX-M-15; 4 isolates coharboring blaCTX-M-14 and blaCTX-M-15; and 3 isolates with blaCTX-M-14, blaCTX-M-3, or blaCTX-M-65. The other acquired β-lactam resistance determinants encoded for the narrow-spectrum β-lactamases OXA-9 and TEM-1. To decipher quinolone resistance, we analyzed the presence of plasmid-mediated resistance determinants and known mutations in gyrase and topoisomerases (20–22). Resistance to quinolones was mediated by either mutation in gyrase gyrA affecting the residues S83 (p.S83I, n = 42; p.S83F, n = 7; and p.S83Y, n = 2) or D87 (p.D87N, n = 7; p.D87G, n = 3; and p.D87A, n = 2) or parC affecting the residues p.S80 (p.S80I, n = 51) or the production of Qnr (Qnr66-like, QnrB6, or QnrS1). Aminoglycoside resistance was caused by the production of the 16S RNA methylase RmtB (n = 7) or an aminoglycoside-modifying enzyme (encoding by aac(3′)-IIa, aadA1, aadA2, aac(6′)-Ib-cr, or strA/strB). Colistin resistance (n = 5) resulted systematically in chromosome-encoded resistance with alteration of the mgrB gene. In these isolates, 2 possessed a nonsense substitution (p.Q30*), 1 missense involved in colistin resistance (p.C27W) (23), an ISKpn26-like inserted (with direct repeats [DRs] of 4 bp: TTAA), and a missense mutation leading to the disappearance of the start codon. No isolate had plasmid-encoded resistance mcr-1.
Genetic Diversity of KPC-Kp
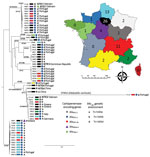
Phylogenetic analysis revealed that the 63 KPC-Kp belonged to 15 different clones (STs) circulating in France (Figure 2; Appendix 1). Although many studies have asserted that CG258 is responsible for the spread of blaKPC (6,8–11), in our collection, only 8 isolates (12.7%) belonged to CG258 (4 each for ST258 and ST512). Furthermore, epidemiologic investigations revealed no link between these isolates (Figure 2). Because KPC is not widely disseminated in France, we did not expect to observe such clonal diversity. Indeed, the epidemiology of KPC in France is not comparable to what was reported in nearby countries in Europe where KPC is endemic, such as Greece and Italy, and where the spread of blaKPC-2/-3 is clearly linked to CG258 (24,25). Among the 8 isolates we identified that belonged to CG258, 3 were recovered from patients with travel history in Greece (isolate 175C3 and isolate 177H5) and Italy (isolate 160C2). In France, the 3 most prevalent clones are ST307 (with 15 isolates), ST147 (12 isolates), and ST13 (7 isolates). By using the 21 SNP cutoff value proposed by David et al. to identity a single hospital outbreak caused by a ST258 and ST512 cluster (8), we identified that the ST307 clone was overrepresented because of an outbreak that included 11 isolates (Figure 2; Appendix 2). However, this ST307 also included 4 isolates that were not related to this outbreak, such as the 195I4 strain, which was isolated from a patient who traveled in Crete (Greece) and possesses an additional carbapenemase-encoding gene (blaNDM-4). The second most prevalent clone, ST147, seemed to have disseminated upon distinct events (Appendix 2). Most of the ST147 isolates have been recovered from different areas with no epidemiologic link between the patients (Figure 2). A link with Portugal has been identified for most (9/11) patients infected or colonized with a KPC-3–producing K. pneumoniae of ST147 (Figure 2). The same link with Portugal was observed for 4 patients infected or colonized with a KPC-3–producing K. pneumoniae of ST231. Strains from ST383 represented a small outbreak for which cross contamination was evidenced (<20 SNPs between 4 isolates [Appendix 2]). One isolate (171J7) was distantly related to other clones and corresponded to K. variicola. KPC-2-producing K. pneumoniae isolates of ST11 were predominantly linked to patients who had a history of travel in Asia (China and Vietnam), where this ST is known to be the main vector of blaKPC dissemination.
Diversity of Genetic Vehicle Involved in Spread of blaKPC
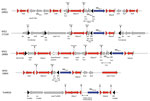
Analysis of the close genetic context of blaKPC highlighted diversity in the genetic structures at the origin of the acquisition of the carbapenemase-encoding gene. The well-known Tn4401a (in 29 isolates) and Tn4401d (in 26 isolates) were the most prevalent structures identified (Figure 2 and 3; Appendix 1). The KPC-Kp of the 2 main clones ST307 and ST147, blaKPC, is carried on Tn4401a in ST307 and Tn4401d in ST147. Two unrelated isolates (ST11–167I9 and the K. variicola 171J7 isolate) harbored blaKPC in the Tn4401b isoform. In the remaining 6 isolates (ST273–171J9, ST147–199D1, ST1788–189B3, ST11–171G8, ST11–190F6, and ST11–171J10), blaKPC-2 is localized in an NTE element (Figures 2, 3). Although 3 isolates belonged to ST11, they displayed 200–800 SNPs of differences along their core genome, indicating that they were unrelated (Appendix 2). The links with 3 different countries (Portugal for ST11–171G8, China for ST11–190F6, and Vietnam for ST11–171J10) are consistent with this unrelatedness. Analysis of NTE elements revealed 4 different structures even if common features were observed (Figure 3). For instance, the presence of a fragment of ISKpn6 downstream of blaKPC and a copy of ISKpn27 upstream were always present (Figure 3). DRs of TATAGG bracketing ISKpn27 indicated a transposition process that occurred inside the resolvase gene of Tn3. Immediately upstream of blaKPC-2 (74 bp), the presence of the inverted repeat right of Tn3 is present in all NTE, indicating that all these structures were related. However, the NTE differed by the size of the deletions that are present between ISKpn27 and blaKPC-2 (from 280 bp in NTE-190F6 to 940 bp in NTE-199D1). We could observe a remnant of blaTEM-1 in longer structures, but it was not functional anymore. Analysis of the 4 NTE revealed that in NTE-199D1, several copies of IS26 bracketed the whole structure, indicating that this IS might be involved in its acquisition by transposition or a recombination event. IS26 has been recently demonstrated to be able to transpose and thus create a class I transposon by targeting another copy of IS26 (26). NTE-171J10 is inserted in the fip gene of IncN-type plasmids with the presence of DRs surrounding the NTE-171J0 (Figure 3). The fip gene has already been demonstrated to be an integration hot spot in IncN-type plasmids (27,28). DRs as well as putative inverted repeats of Tn3-family transposon are present at the integration site (Figure 3). Moreover, the presence of the complete Tn3 transposase gene indicated that NTE-171J10 might be functional. In NTE-189B3, a new class I transposon carrying a protein of unknown function has been identified. DRs bracketed ISApu1 and ISApu2, indicating a transposition process mediated by these close insertion sequences (Figure 3).
In France, KPC producers (84.1% of K. pneumoniae) represent only 6.8% of all carbapenemase producers, far away from the global 72.7% found in Europe in 2013 (8). This relatively low prevalence of KPC producers in France compared with OXA-48–like and NDM producers has been reported since 2012 (3,4,29). We demonstrated unexpected clonal diversity among KPC-Kp isolated in France. A few overrepresented clones were identified (i.e., ST307, ST147, ST231, and ST13). However, ST307 was involved in a regional outbreak, whereas ST147 and ST13 were identified in different parts of France. Most of the patients colonized or infected with KPC-Kp had a clear link with Portugal, where these 4 STs were recently described to be the more prevalent (30,31). The KPC-2–producing K. pneumoniae isolates identified in France were predominantly recovered from patients with a history of travel in Greece (ST258) or Asia (ST11).
Regarding antimicrobial susceptibility of KPC-Kp in France, the relative high susceptibility to imipenem (30.3%) and meropenem to a lesser extent (18.2%) are in agreement with previous reports from Italy, where ST512 is highly prevalent (26.6% susceptibility to meropenem) (32). Conversely, data from the United States and Taiwan indicated that KPC-Kp are more resistant to carbapenems in those parts of the world, where ST258 is more prevalent (33,34).
Altogether, our results indicate that the KPC-Kp epidemiology has changed in Europe during the past 5 years. In 2018, ST258 and ST512 K. pneumoniae were no longer the main drivers of KPC resistance, at least in several non–KPC-endemic countries of western Europe, such as France and Portugal (30,31). KPC-Kp epidemiology also appears to have begun changing in some countries, such as Italy and Colombia, where CG-258 KPC-Kp was previously known to be endemic. This change is indicated by the reported emergence of ST307 and ST273 KPC-Kp in Sicilia (Italy) (35) and ST307 and ST14 KPC-Kp in Colombia (36). This change in the global epidemiology of KPC-Kp might have an effect on the identification of these carbapenemase producers with the molecular methods dedicated to the identification of GC258 K. pneumoniae (37,38).
In addition, our study highlights the dissemination of blaKPC genes in high-risk clones of K. pneumoniae (ST307 and ST147), genetic features that might provide an advantage in adaptation to the hospital environment and the human host (39). These clones already convey several antimicrobial-resistance genes, including genes encoding other carbapenemases of NDM and OXA-48–like types (40,41). Accordingly, we might now fear the emergence of ST307 and ST147 high-risk clones of K. pneumoniae that can co-produce multiple carbapenemases. A recent study demonstrated the importance of ST307 in the dissemination of blaOXA-181 in South Africa (42). In that study, >600 isolates belonging to ST307 were recovered and analyzed, and the results demonstrated the importance of this clone as a carrier of carbapenemase genes in all continents. Another study used Bayesian analysis to demonstrate that ST307 emerged in the mid-1990s (43). ST307 had been strongly associated with the diffusion of blaCTX-M-15 (43) and now is associated with the dissemination of carbapenemase genes (42).
In conclusion, we found that the epidemiology of KPC-Kp has changed in Europe, in particular, with emergence of non–CG258 KPC-Kp isolates in France, linked to dissemination from Portugal. This change in epidemiology has to be considered by microbiologists because a few diagnostic assays specifically designed for the identification of ST-258 KPC-Kp isolates will not be able to detect non–CG258 KPC-Kp isolates.
Dr. Bonnin is an associate professor in Medical Microbiology at the University of Paris-Sud Medical School and a member of the Associated National Reference Center for Carbapenemase-Producing Enterobacteriaceae. His research focuses on bacterial genomes and transcriptomes with a special interest in mobile genetic elements and evolution.
Acknowledgment
This work was funded in part by the University Paris-Sud, France. L.D., T.N., and R.A.B. are members of the Laboratory of Excellence in Research on Medication and Innovative Therapeutics, which is supported by a grant from France’s National Research Agency (grant no. ANR-10-LABX-33).
References
- Munoz-Price LS, Poirel L, Bonomo RA, Schwaber MJ, Daikos GL, Cormican M, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13:785–96. DOIPubMedGoogle Scholar
- Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25:682–707. DOIPubMedGoogle Scholar
- Dortet L, Cuzon G, Nordmann P. Dissemination of carbapenemase-producing Enterobacteriaceae in France, 2012. J Antimicrob Chemother. 2014;69:623–7. DOIPubMedGoogle Scholar
- Dortet L, Cuzon G, Ponties V, Nordmann P. Trends in carbapenemase-producing Enterobacteriaceae, France, 2012 to 2014. Euro Surveill. 2017;22:22. DOIPubMedGoogle Scholar
- Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–36. DOIPubMedGoogle Scholar
- Cuzon G, Naas T, Truong H, Villegas MV, Wisell KT, Carmeli Y, et al. Worldwide diversity of Klebsiella pneumoniae that produce β-lactamase blaKPC-2 gene. Emerg Infect Dis. 2010;16:1349–56. DOIPubMedGoogle Scholar
- Fu P, Tang Y, Li G, Yu L, Wang Y, Jiang X. Pandemic spread of bla KPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int J Antimicrob Agents. 2019;54:117–24. DOIPubMedGoogle Scholar
- David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, et al.; EuSCAPE Working Group; ESGEM Study Group. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol. 2019;4:1919–29. DOIPubMedGoogle Scholar
- Chmelnitsky I, Shklyar M, Hermesh O, Navon-Venezia S, Edgar R, Carmeli Y. Unique genes identified in the epidemic extremely drug-resistant KPC-producing Klebsiella pneumoniae sequence type 258. J Antimicrob Chemother. 2013;68:74–83. DOIPubMedGoogle Scholar
- Fortini D, Villa L, Feudi C, Pires J, Bonura C, Mammina C, et al. Double copies of bla(KPC-3):Tn4401a on an IncX3 plasmid in Klebsiella pneumoniae successful clone ST512 from Italy. Antimicrob Agents Chemother. 2015;60:646–9. DOIPubMedGoogle Scholar
- Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53:3365–70. DOIPubMedGoogle Scholar
- Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P. Genetic structures at the origin of acquisition of the β-lactamase bla KPC gene. Antimicrob Agents Chemother. 2008;52:1257–63. DOIPubMedGoogle Scholar
- Araújo BF, Royer S, Campos PA, Ferreira ML, Gonçalves IR, Machado LG, et al. Insights into a novel Tn4401 deletion (Tn4401i) in a multidrug-resistant Klebsiella pneumoniae clinical strain belonging to the high-risk clonal group 258 producing KPC-2. Int J Antimicrob Agents. 2018;52:525–7. DOIPubMedGoogle Scholar
- Naas T, Cuzon G, Truong HV, Nordmann P. Role of ISKpn7 and deletions in blaKPC gene expression. Antimicrob Agents Chemother. 2012;56:4753–9. DOIPubMedGoogle Scholar
- Girlich D, Bonnin RA, Jousset A, Naas T. Promoter characterization and expression of the blaKPC-2 gene in Escherichia coli, Pseudomonas aeruginosa and Acinetobacter baumannii. J Antimicrob Chemother. 2017;72:1597–601. DOIPubMedGoogle Scholar
- Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–96. DOIPubMedGoogle Scholar
- Girlich D, Bonnin RA, Bogaerts P, De Laveleye M, Huang DT, Dortet L, et al. Chromosomal amplification of the blaOXA-58 carbapenemase gene in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother. 2017;61:61. DOIPubMedGoogle Scholar
- Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. DOIPubMedGoogle Scholar
- Jousset AB, Oueslati S, Creton E, Cotellon E, Sauvadet A, Emeraud C, et al. Resistance to ceftazidime-avibactam in a Klebsiella pneumoniae clinical isolate due to the production of KPC-39, a single amino-acid variant of KPC-3. Presented at: 29th European Congress of Clinical Microbiology and Infectious Diseases; Amsterdam, the Netherlands; 2019 April 13–16.
- Ruiz J, Goñi P, Marco F, Gallardo F, Mirelis B, Jimenez De Anta T, et al. Increased resistance to quinolones in Campylobacter jejuni: a genetic analysis of gyrA gene mutations in quinolone-resistant clinical isolates. Microbiol Immunol. 1998;42:223–6. DOIPubMedGoogle Scholar
- Vila J, Ruiz J, Goñi P, De Anta MT. Detection of mutations in parC in quinolone-resistant clinical isolates of Escherichia coli. Antimicrob Agents Chemother. 1996;40:491–3. DOIPubMedGoogle Scholar
- Vila J, Ruiz J, Marco F, Barcelo A, Goñi P, Giralt E, et al. Association between double mutation in gyrA gene of ciprofloxacin-resistant clinical isolates of Escherichia coli and MICs. Antimicrob Agents Chemother. 1994;38:2477–9. DOIPubMedGoogle Scholar
- Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30:557–96. DOIPubMedGoogle Scholar
- Conte V, Monaco M, Giani T, D’Ancona F, Moro ML, Arena F, et al.; AR-ISS Study Group on Carbapenemase-Producing K. pneumoniae. Molecular epidemiology of KPC-producing Klebsiella pneumoniae from invasive infections in Italy: increasing diversity with predominance of the ST512 clade II sublineage. J Antimicrob Chemother. 2016;71:3386–91. DOIPubMedGoogle Scholar
- Gartzonika K, Rossen JWA, Sakkas H, Rosema S, Priavali E, Friedrich AW, et al. Identification of a KPC-9-producing Klebsiella pneumoniae ST258 cluster among KPC-2-producing isolates of an ongoing outbreak in Northwestern Greece: a retrospective study. Clin Microbiol Infect. 2018;24:558–60. DOIPubMedGoogle Scholar
- Harmer CJ, Hall RM. IS26-mediated formation of transposons carrying antibiotic resistance genes. MSphere. 2016;1:1. DOIPubMedGoogle Scholar
- Gauthier L, Dortet L, Jousset AB, Mihaila L, Golse N, Naas T, et al. Molecular characterization of plasmid-encoded Tripoli MBL 1 (TMB-1) in Enterobacteriaceae. J Antimicrob Chemother. 2019;74:42–7.PubMedGoogle Scholar
- Poirel L, Bonnin RA, Nordmann P. Analysis of the resistome of a multidrug-resistant NDM-1-producing Escherichia coli strain by high-throughput genome sequencing. Antimicrob Agents Chemother. 2011;55:4224–9. DOIPubMedGoogle Scholar
- Lee CR, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol. 2016;7:895. DOIPubMedGoogle Scholar
- Aires-de-Sousa M, Ortiz de la Rosa JM, Gonçalves ML, Pereira AL, Nordmann P, Poirel L. Epidemiology of carbapenemase-producing Klebsiella pneumoniae in a hospital, Portugal. Emerg Infect Dis. 2019;25:1632–8. DOIPubMedGoogle Scholar
- Rodrigues C, Bavlovič J, Machado E, Amorim J, Peixe L, Novais Â. KPC-3-producing Klebsiella pneumoniae in Portugal linked to previously circulating non-CG258 lineages and uncommon genetic platforms (Tn4401d-IncFIA and Tn4401d-IncN). Front Microbiol. 2016;7:1000. DOIPubMedGoogle Scholar
- Cojutti P, Sartor A, Bassetti M, Scarparo C, Pea F. Is meropenem MIC increase against KPC-producing Klebsiella pneumoniae correlated with increased resistance rates against other antimicrobials with Gram-negative activity? J Glob Antimicrob Resist. 2018;14:238–41. DOIPubMedGoogle Scholar
- Chiu SK, Ma L, Chan MC, Lin YT, Fung CP, Wu TL, et al. Carbapenem nonsusceptible Klebsiella pneumoniae in Taiwan: dissemination and increasing resistance of carbapenemase producers during 2012–2015. Sci Rep. 2018;8:8468. DOIPubMedGoogle Scholar
- Kaiser RM, Castanheira M, Jones RN, Tenover F, Lynfield R. Trends in Klebsiella pneumoniae carbapenemase-positive K. pneumoniae in US hospitals: report from the 2007-2009 SENTRY Antimicrobial Surveillance Program. Diagn Microbiol Infect Dis. 2013;76:356–60. DOIPubMedGoogle Scholar
- Bonura C, Giuffrè M, Aleo A, Fasciana T, Di Bernardo F, Stampone T, et al.; MDR-GN Working Group. An update of the evolving epidemic of blaKPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple non-ST258 clones. PLoS One. 2015;10:
e0132936 . DOIPubMedGoogle Scholar - Ocampo AM, Chen L, Cienfuegos AV, Roncancio G, Chavda KD, Kreiswirth BN, et al. A two-year surveillance in five Colombian tertiary care hospitals reveals high frequency of non-CG258 clones of carbapenem-resistant Klebsiella pneumoniae with distinct clinical characteristics. Antimicrob Agents Chemother. 2015;60:332–42. DOIPubMedGoogle Scholar
- Chen L, Chavda KD, Mediavilla JR, Zhao Y, Fraimow HS, Jenkins SG, et al. Multiplex real-time PCR for detection of an epidemic KPC-producing Klebsiella pneumoniae ST258 clone. Antimicrob Agents Chemother. 2012;56:3444–7. DOIPubMedGoogle Scholar
- Yu F, Lv J, Niu S, Du H, Tang YW, Pitout JDD, et al. Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (sequence type 258 [ST258] and ST11) and hypervirulent (ST23, ST65, ST86, and ST375) strains. J Clin Microbiol. 2018;56:56. DOIPubMedGoogle Scholar
- Villa L, Feudi C, Fortini D, Brisse S, Passet V, Bonura C, et al. Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb Genom. 2017;3:
e000110 . DOIPubMedGoogle Scholar - Potel C, Ortega A, Martínez-Lamas L, Bautista V, Regueiro B, Oteo J. Interspecies transmission of the blaOXA-48 gene from a Klebsiella pneumoniae high-risk clone of sequence type 147 to different Escherichia coli clones in the gut microbiota. Antimicrob Agents Chemother. 2017;62:62. DOIPubMedGoogle Scholar
- Turton J, Davies F, Turton J, Perry C, Payne Z, Pike R. Hybrid resistance and virulence plasmids in “high-risk” clones of Klebsiella pneumoniae, including those carrying blaNDM-5. Microorganisms. 2019;7:7. DOIPubMedGoogle Scholar
- Lowe M, Kock MM, Coetzee J, Hoosien E, Peirano G, Strydom K-A, et al. Klebsiella pneumoniae ST307 with blaOXA-181, South Africa, 2014-2016. Emerg Infect Dis. 2019;25:739–47. DOIPubMedGoogle Scholar
- Wyres KL, Hawkey J, Hetland MAK, Fostervold A, Wick RR, Judd LM, et al. Emergence and rapid global dissemination of CTX-M-15-associated Klebsiella pneumoniae strain ST307. J Antimicrob Chemother. 2019;74:577–81. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: May 06, 2020
Table of Contents – Volume 26, Number 6—June 2020
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Laurent Dortet, Service de Bactériologie-Hygiène, Hôpital de Bicêtre, 78 rue du Général Leclerc, 94275 Le Kremlin-Bicêtre CEDEX, France
Top