Volume 26, Number 7—July 2020
Dispatch
Shuni Virus in Wildlife and Nonequine Domestic Animals, South Africa
Figure 2
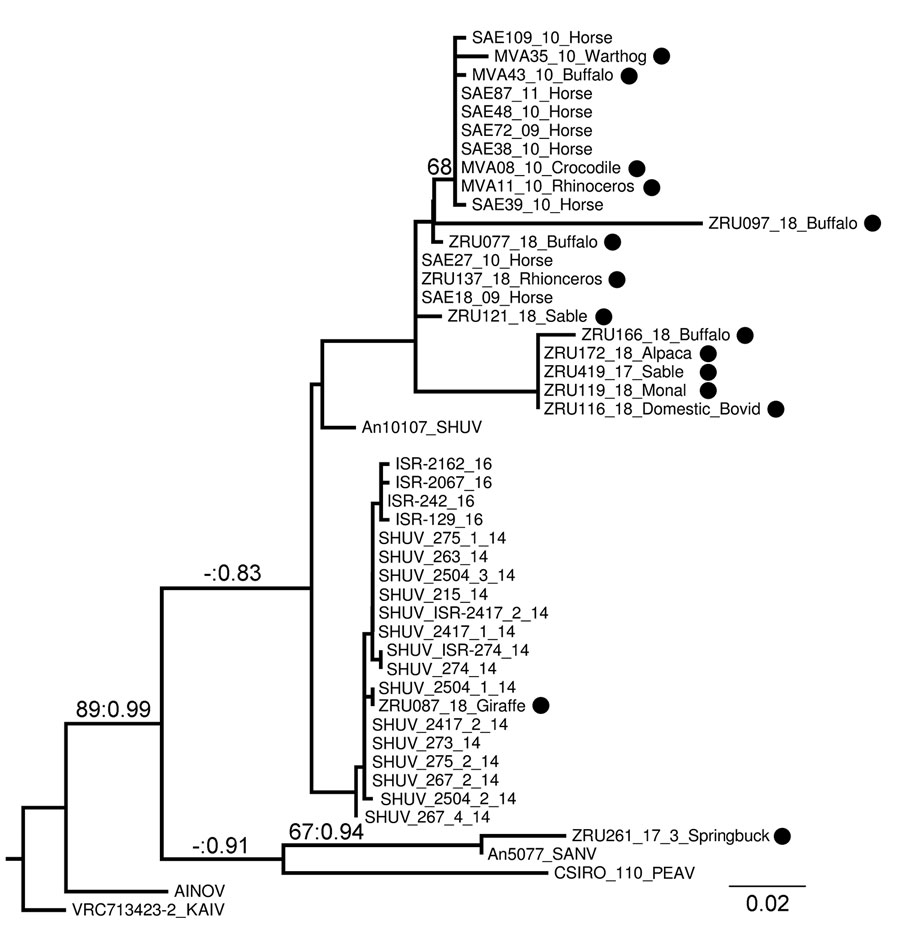
Figure 2. Phylogram of clade 1a, lineage I, of the Simbu serogroup (15) recovered from maximum-likelihood and Bayesian analyses of the small segment for SHUV isolates from wildlife and nonequine domestic animals, South Africa and reference sequences. Bootstrap values (maximum likelihood >60) and posterior probabilities (>0.8) are displayed on branches as support values. GenBank accession numbers for sequences from this study (black circles): MVA11_10_Rhinoceros, JQ726395; MVA08_10_Crocodile, JQ726396; MVA43_10_Buffalo, JQ726397; MVA35_10_Warthog, JQ726398; ZRU077_18_Buffalo, MK114084; ZRU121_18_Sable, MK114085; ZRU137_18_Rhinoceros, MK114086. GenBank accession numbers, virus types, and locations for reference sequences: An10107, AF362405, SHUV Nigeria; AINOV, M22011, Japan; VRC713423–2 KAIV, AF362394, India; SAE72_09_Horse, HQ610138, South Africa; SAE27_10_Horse, HQ610139, South Africa; SAE38_10_Horse, HQ610140, South Africa; SAE39_10_Horse, HQ610141, South Africa; SAE48_10_Horse, HQ610142, South Africa; SAE109_10_Horse, HQ610143, South Africa; SAE18_09_Horse, KC510272, South Africa; SAE87_11_Horse, KC525997, South Africa; Shuni_215_14, KP900859, Israel; Shuni_263_14, KP900860, Israel; Shuni_267_2_14, KP900861, Israel; Shuni_267_4_14, KP900862, Israel; Shuni_273_14, KP900865, Israel; Shuni_274_14, KP900867, Israel; Shuni_275_1_14, KP900869, Israel; Shuni_275_2_14, KP900871, Israel; Shuni_2417_1_14, KP900872, Israel; Shuni_2417_2_14, KP900875, Israel; Shuni_2504_1_14, KP900877, Israel; Shuni_2504_2_14, KP900878, Israel; Shuni_2504_3_14, KP900882, Israel; 2504_3_14, KU937313, Israel; SHUV_ISR-274_14, KT946779, Israel; SHUV_ISR-2417_2_14, KT946780, Israel; ISR-129_16, MF361846, Israel; ISR-242_16, MF361849, Israel; ISR-2067_16, MF361852, Israel; ISR-2162_16, MF361855, Israel; CSIRO 110, MH484320, Australia; An5077, AF362402, Nigeria. AINOV, ainovirus; KAIV, kaikalurvirus; PEAV, Peaton virus; SANV, Sango virus; SHUV, Shuni virus..