Volume 27, Number 1—January 2021
Dispatch
Limited Specificity of Serologic Tests for SARS-CoV-2 Antibody Detection, Benin
Cite This Article
Citation for Media
Abstract
We used commercially available ELISAs to test 68 samples from coronavirus disease cases and prepandemic controls from Benin. We noted <25% false-positive results among controls, likely due to unspecific immune responses elicited by acute malaria. Serologic tests must be carefully evaluated to assess coronavirus disease spread and immunity in tropical regions.
Since its emergence in China late 2019, coronavirus disease (COVID-19) had caused >41 million cases and >1.1 million deaths globally by October 2020, according to the World Health Organization (https://www.who.int/publications/m/item/weekly-operational-update---30-october-2020). Diagnosis of the causative pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is based on reverse transcription-PCR (RT-PCR) to detect viral nucleic acid or serologic assays to detect SARS-CoV-2 antigens in early stages of disease (1,2). In later stages of disease, antibody-based serologic testing can complement diagnosis of SARS-CoV-2 infection. In addition, antibody-based serologic testing is a valuable epidemiologic tool to assess COVID-19 spread and potential immunity to SARS-CoV-2. Serologic studies in Europe and Asia indicate high sensitivity and specificity of widely used SARS-CoV-2 antibody ELISAs (3,4). However, many serologic tests have not been validated in resource-limited settings (5). We conducted a SARS-CoV-2 serologic assessment in Benin by using samples from patients with RT-PCR–confirmed SARS-CoV-2 infection and controls sampled before the first SARS-CoV-2 detection in March 2020.
We obtained convalescent serum samples from 8 patients in Benin with RT-PCR–confirmed COVID-19 during March–April 2020. The average sampling time was 8 (range 1–10) days after RT-PCR confirmation of SARS-CoV-2 infection (Table 1). We also included 60 serum samples from patients with acute febrile illness tested as part of hemorrhagic fever surveillance during October–November 2019 as prepandemic controls (Table 2). Sampling was approved by the ethics committee of the Benin Ministry of Health (approval no. 030/MS/DC/SGM/DNSP/CJ/SA/027SGG2020).
We tested all 68 serum samples by using commercially available ELISAs from EUROIMMUN (https://www.euroimmun.com) that rely on different antigens and antibody classes: SARS-CoV-2 nucleocapsid (N) antigen (IgG), spike 1 (S1) subunit (IgG and IgA), and Middle East respiratory syndrome coronavirus (MERS-CoV) S1 (IgG). We also used the SCoV-2 Detect IgG ELISA (InBios, https://inbios.com), an IgG-only S1 antigen-based test authorized for emergency use by the US Food and Drug Administration. Serum samples also were tested by using commercially available ELISA kits (Euroimmun) against the Zika virus (ZIKV) nonstructural protein 1 (NS1) antigen (IgG), the Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA1) (IgG), and the EBV viral capsid (CA) antigen (IgM and IgG), as well as real-time PCR tests (TIB MOLBIOL, https://www.tib-molbiol.com) for all human pathogenic Plasmodium species, EBV, and cytomegalovirus (CMV). Plaque-reduction neutralization tests (PRNTs) were performed by using similar methods for SARS-CoV-2 and ZIKV as described (4,6). We used previously described recombinant S-based immunofluorescence assays (7) to test for specific antibodies to common cold betacoronavirus human coronavirus (HCoV) OC43 and HCoV-HKU1.
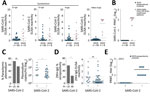
Among the 8 patients with RT-PCR–confirmed SARS-CoV-2 infection, seroconversion ranged from 62.5%–100% (95% CI 30.8%–100.0%), depending on the ELISA used (Figure 1, panel A), suggesting differential sensitivity of ELISAs on the basis of immunoglobulin detected and the commercial kit used. Indeed, early after infection, IgA-based tests had a higher sensitivity than most IgG-based SARS-CoV-2 ELISAs; only the InBios IgG-based kit was positive for all RT-PCR–confirmed patients (Figure 1, panel A). A total of 87.5% (7/8) of ELISA results were confirmed by a highly specific SARS-CoV-2 PRNT (Figure 1, panel B).
When summarizing all antibody classes, antigens, and kits among the 60 prepandemic controls, we observed 25.0% (15/60; 95% CI 15.7%–37.3%) positive or borderline ELISA results (8). Different from RT-PCR–confirmed cases, ELISA reactivity in those samples contrasted with the complete lack of SARS-CoV-2–specific neutralizing antibodies, suggesting unspecific ELISA reactivity (Figure 1, panel B).
Unspecific SARS-CoV-2 ELISA reactivity might be consistent with, but not limited to, 3 scenarios. First, antibodies elicited by common infections with endemic human coronaviruses might cross-react with SARS-CoV-2 antigens (1). However, a Fisher exact test showed no statistically significant difference in the frequency of antibody reactivity with common cold coronavirus antigens between SARS-CoV-2 ELISA-positive serum samples compared with SARS-CoV-2 ELISA-negative samples. In detail, reactivity with HCoV-OC43 was 63.6% in SARS-CoV-2 ELISA-positive samples and 70.4% in SARS-CoV-2 ELISA-negative samples (p = 0.7); reactivity with HCoV-HKU-1 was 45.7% in SARS-CoV-2 ELISA-positive samples and 74.0% in SARS-CoV-2 ELISA-negative samples (p = 0.1) (Appendix Figure 1, panel A). Similarly, a Student t-test revealed no statistically significant difference in the magnitude of antibody titers against common cold coronaviruses between SARS-CoV-2 ELISA-positive or ELISA-negative samples (p = 0.09 for HCoV-OC43 and p = 0.8 for HCoV-HKU1) (Appendix Figure 1, panel B). Of note, no serum reacted with MERS-CoV antigens, suggesting that unspecific reactivity might not apply to all coronavirus antigens and tests (Appendix Figure 2). Second, polyclonal B-cell activation can occur in infections with or reactivations of herpesviruses, such as CMV and EBV, and elicit false-positive results in serologic tests (9). However, only 2 patients had a positive CMV PCR and only 1 patient had a positive EBV PCR (Figure 2). In addition, persons with SARS-CoV-2 ELISA-positive versus ELISA-negative results did not differ in their past exposure to EBV, according to detailed serologic analyses (Figure 2; Appendix Figure 3). Finally, polyclonal B cell activation also can be caused by acute malaria, which is widespread in Africa (10). More (71.4%) persons with SARS-CoV-2–positive ELISAs than those with negative ELISAs (54.3%) were positive for Plasmodium in a highly sensitive PCR test, but the difference was not statistically significant by Fisher exact test (p = 0.35; Figure 1, panel C). However, parasite loads were statistically significantly higher among SARS-CoV-2 ELISA-positive than ELISA-negative persons by Student t-test (p = 0.035; Figure 1, panel C). In malaria, higher parasite loads are detected at early stages of infection and decrease over time, suggesting a higher proportion of acute malaria in SARS-CoV-2 ELISA–positive patients compared with likely subacute or chronic malaria in SARS-CoV-2 ELISA–negative patients (11). Thus, acute malaria is the most plausible explanation for unspecific SARS-CoV-2 ELISA reactivity in prepandemic controls. To assess the breadth of unspecific reactivity, we tested the serum samples from prepandemic controls by using a ZIKV IgG ELISA, for which unspecific reactivity has been reported in cases of acute malaria (10). We found that 57.1% of samples that elicited potentially unspecific SARS-CoV-2 ELISA results also showed ZIKV ELISA–positive results, whereas only 23.9% of samples that were SARS-CoV-2 ELISA–negative were ZIKV ELISA–positive. This difference was statistically significant by Fisher exact test (p = 0.019) (Figure 1, panel D; Appendix Figure 4). From the prepandemic controls that were SARS-CoV-2 ELISA positive, no ZIKV ELISA–positive serum samples showed ZIKV-specific neutralizing antibodies, suggesting unspecific reactivity of those samples in the ZIKV ELISA, similar to the discrepant results of SARS-CoV-2 ELISA and PRNT observed in those serum samples (Figure 1, panel E; Figure 2).
We assessed SARS-CoV-2 antibody-based serologic diagnostics in Benin and noted unspecific reactivity in up to 25% of febrile patients, possibly due to acute malaria. Limitations of our study include the small sample size and limited patient metadata. Testing of serum samples for CMV and EBV by PCR might not have been sensitive due to lack of cell-associated viral nucleic acid; therefore, we cannot exclude potential herpesvirus reactivation affecting serologic testing. Nevertheless, our analyses point to acute malaria as the likely cause of the unspecific serologic reactivity, although we cannot exclude other coexisting conditions in the tropics, such as dengue virus, which also can affect testing (12).
Unspecific reactivity in serologic tests might affect public health interventions in tropical regions, leading to overestimates of SARS-CoV-2 circulation in regions where malaria is endemic and to misidentification of SARS-CoV-2 hotspots. In addition, due to false-positive SARS-CoV-2 results, target populations for vaccine campaigns might be missed when vaccines become available, and coexistent diseases, such as malaria, might be overlooked, leading to higher mortality rates from endemic diseases (13,14). The robustness of current and future SARS-CoV-2 serologic tests should be further assessed by multicentric seroepidemiologic studies from different tropical regions (15).
Dr. Yadouleton is a medical entomologist in the Centre de Recherche Entomologique de Cotonou, Benin, head of the Laboratoire des Fièvres Hémorragiques in Cotonou, and a teacher at the University of Natitingou, Benin. His research interests include mosquito control and the diagnosis of viral hemorrhagic fevers.
Acknowledgment
This article was preprinted at https://www.medrxiv.org/content/10.1101/2020.06.29.20140749v1.
References
- Meyer B, Drosten C, Müller MA. Serological assays for emerging coronaviruses: challenges and pitfalls. Virus Res. 2014;194:175–83. DOIPubMedGoogle Scholar
- He X, Lau EHY, Wu P, Deng X, Wang J, Hao X, et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat Med. 2020;26:672–5. DOIPubMedGoogle Scholar
- Zhao R, Li M, Song H, Chen J, Ren W, Feng Y, et al. Early detection of SARS-CoV-2 antibodies in COVID-19 patients as a serologic marker of infection. Clin Infect Dis. 2020;ciaa523.
- Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease 2019 patients. Emerg Infect Dis. 2020;26:1478–88. DOIPubMedGoogle Scholar
- Fischer C, Drosten C, Drexler JF. The difficulties in obtaining reliable Zika virus diagnostics. Lancet Infect Dis. 2019;19:240–1. DOIPubMedGoogle Scholar
- Netto EM, Moreira-Soto A, Pedroso C, Höser C, Funk S, Kucharski AJ, et al. High Zika virus seroprevalence in Salvador, northeastern Brazil limits the potential for further outbreaks. MBio. 2017;8:e01390–17. DOIPubMedGoogle Scholar
- Corman VM, Müller MA, Costabel U, Timm J, Binger T, Meyer B, et al. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17:20334. DOIPubMedGoogle Scholar
- Harvey R, Mattiuzzo G, Hassall M, Sieberg A, Müller MA, Drosten C, et al.; study participants. study participants. Comparison of serologic assays for Middle East respiratory syndrome coronavirus. Emerg Infect Dis. 2019;25:1878–83. DOIPubMedGoogle Scholar
- Sangster MY, Topham DJ, D’Costa S, Cardin RD, Marion TN, Myers LK, et al. Analysis of the virus-specific and nonspecific B cell response to a persistent B-lymphotropic gammaherpesvirus. J Immunol. 2000;164:1820–8. DOIPubMedGoogle Scholar
- Van Esbroeck M, Meersman K, Michiels J, Arien KK, Van den Bossche D. Letter to the editor: Specificity of Zika virus ELISA: interference with malaria. Euro Surveill. 2016;21(21).
- Dormond L, Jaton-Ogay K, de Vallière S, Genton B, Bille J, Greub G. Multiplex real-time PCR for the diagnosis of malaria: correlation with microscopy. Clin Microbiol Infect. 2011;17:469–75. DOIPubMedGoogle Scholar
- Lustig Y, Keler S, Kolodny R, Ben-Tal N, Atias-Varon D, Shlush E, et al. Potential antigenic cross-reactivity between SARS-CoV-2 and Dengue viruses. Clin Infect Dis. 2020;
ciaa1207 ; Epub ahead of print. DOIPubMedGoogle Scholar - Fischer C, de Oliveira-Filho EF, Drexler JF. Viral emergence and immune interplay in flavivirus vaccines. Lancet Infect Dis. 2020;20:15–7. DOIPubMedGoogle Scholar
- Plucinski MM, Guilavogui T, Sidikiba S, Diakité N, Diakité S, Dioubaté M, et al. Effect of the Ebola-virus-disease epidemic on malaria case management in Guinea, 2014: a cross-sectional survey of health facilities. Lancet Infect Dis. 2015;15:1017–23. DOIPubMedGoogle Scholar
- Elm J, Desowitz R, Diwan A. Serological cross-reactivities between the retroviruses HIV and HTLV-1 and the malaria parasite Plasmodium falciparum. P N G Med J. 1998;41:15–22.PubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: December 01, 2020
1These first authors contributed equally to this article.
2These senior authors contributed equally to this article.
Table of Contents – Volume 27, Number 1—January 2021
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Jan Felix Drexler, Helmut-Ruska-Haus, Institute of Virology, Campus Charité Mitte, Charitéplatz 1, Berlin 10117, Germany
Top