Volume 27, Number 12—December 2021
Dispatch
Detection of SARS-CoV-2 in Wastewater at Residential College, Maine, USA, August–November 2020
Abstract
We used wastewater surveillance to identify 2 coronavirus disease outbreaks at a college in Maine, USA. Cumulative increases of >1 log10 severe acute respiratory syndrome coronavirus 2 RNA in consecutive 24-hour composite samples preceded the outbreaks. For 76% of cases, RNA was identified in grab samples from residence halls <7 days before case discovery.
Wastewater surveillance can indicate the presence and temporal trends of coronavirus disease (COVID-19) cases in a sewershed (1,2). Large universities have used wastewater surveillance to identify residence halls at high risk for transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19 (3,4). We demonstrate that wastewater surveillance using grab samples collected from residential halls and 24-hour composite samples from lift stations can detect COVID-19 outbreaks at a small residential college.
During August 21–November 20, 2020 (days 0–92), we collected weekly grab samples of flowing untreated wastewater from 6 residence halls at a residential college in Maine, USA. The residence halls served 605 students; hall A housed 64 students, hall B housed 127 students, hall C housed 80 students, hall D housed 109 students, hall E housed 87 students, and hall F housed 138 students. During days 13–92 we also collected 24-hour composite samples approximately twice a week from 2 lift stations (i.e., L1 and L3) where wastewater from various buildings on campus was consolidated in holding tanks and pumped to septic tanks; these composite samples represented the total population in the residence halls. L1 contained effluent from halls E–F and L3 contained effluent from halls A–D. Both lift stations also contained effluent from other campus buildings. The wastewater was collected and stored at 4°C for <72 hours before we assayed 105-mL samples using the Water SARS-CoV-2 RT-PCR test (IDEXX Laboratories, Inc., https://www.idexx.com) according to the manufacturer’s instructions. The 2019-nCoV_N_Positive Control plasmid (Integrated DNA Technologies, Inc., https://www.idtdna.com) had a limit of quantification of 2 (average cycle threshold [Ct] 39.42) copies per reaction, and the purified 2019-nCoV_N_Positive Control plasmid cloned into Escherichia coli had a limit of quantification of 20 (average Ct 35.95) copies per reaction. The theoretical limit of detection in all samples was 1 copy per reaction. We included a negative extraction control, no-template control, and a step from the standard curve (2 × 103 or 2 × 104 copies/reaction) in each run. We calculated SARS-CoV-2 RNA concentrations as copies per day per person for 24-hour composite samples and copies per liter per person for grab samples. Nondetectable samples were reported at one half the theoretical limit of detection (5).
Each week, ≈100 students were randomly selected for individual surveillance testing by reverse transcription PCR. We instituted expanded surveillance testing for students living in residence halls with detectable RNA in grab samples or served by lift stations that had increased RNA concentrations.
Affected students were isolated or quarantined according to guidelines from the Centers for Disease Control and Prevention (6,7). COVID-19 outbreak investigations by the Maine Center for Disease Control and Prevention (Augusta, Maine, USA) end 28 days (2 infectious periods) after the specimen collection date of the last identified case. All residential students were tested for SARS-CoV-2 upon outbreak identification, beginning on day 25 for the first outbreak and day 81 for the second outbreak. Widespread testing among students continued weekly until all students had negative test results for 2 successive rounds during the outbreak period.
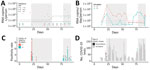
On day 18, we detected SARS-CoV-2 RNA from the wastewater discharge of hall F and both lift stations, preceding an outbreak on days 21–58 (Figure, panels A, B). By day 22, RNA concentrations in both lift stations increased by >1 log10; subsequently, concentrations decreased by >1 log10 by day 33 for L3 and day 40 for L1 (Figure, panel B). On day 21, expanded individual surveillance testing of half of the students living in hall F identified 2 COVID-19 cases (Figure, panels C, D). Widespread surveillance testing beginning on day 22 identified 6 additional cases in hall F, 1 case in hall D, and 1 case in hall E (Figure, panel C). In response, the college implemented remote learning during days 22–36.
Students returned to their residence halls from quarantine or isolation on days 21–45; we observed an increase of SARS-CoV-2 RNA in L3 on days 40–46 (Figure, panel B). We detected RNA in grab samples from hall D on days 45 and 53 and hall F on days 50 and 53 (Figure, panel A); however, concentrations were undetectable in L3 on days 49–70 (Figure, panel B). On day 53, individual surveillance testing of all students in hall F did not identify any COVID-19 cases (Figure, panels C, D).
From days 70 to 82, SARS-CoV-2 RNA concentrations in L3 increased by >1 log10. A second outbreak occurred during days 78–92. Expanded surveillance testing of all students in hall B identified 3 COVID-19 cases, prompting an outbreak investigation. During days 83–89, widespread testing of all students detected 7 additional COVID-19 cases in hall B, 1 case in hall C, and 1 case in hall E (Figure, panel C). The first COVID-19 case in hall E was discovered on day 84, when SARS-CoV-2 RNA was undetectable in L1 (Figure, panel B). The discovery of 7 cases in hall B on day 86 indicated that transmission might be widespread. During days 88–92, the college instituted an exit strategy comprising remote learning and widespread surveillance testing of all students 48 hours before departing campus. On day 91, we detected SARS-CoV-2 RNA in L1 and identified 3 COVID-19 cases (1 in hall A, 1 in hall B, and 1 in hall E; Figure, panels B, C). We detected RNA in grab samples from hall D 4 days before the outbreak as well as during the outbreak and staged dismissal but did not identify any cases in hall D.
We found that a >1 log10 increase of SARS-CoV-2 RNA concentrations in composite samples from a lift station preceded 2 COVID-19 outbreaks at a college campus. RNA concentrations did not substantially increase during the outbreak even as more cases were discovered (Figure, panels B, C). Thus, wastewater surveillance is best used to discover the onset of outbreaks at a small college campus.
We hypothesized that detectable concentrations of SARS-CoV-2 RNA in the wastewater discharge in L1 on days 40–56 (Figure, panel B) were caused by residual shedding among recovering students who were no longer infectious, as described in clinical data (8,9). In addition, a study in Utah, USA, documented evidence of residual shedding in municipal wastewater systems after a decrease in reported cases (2). In L1, SARS-CoV-2 RNA was undetectable 26 days in the wastewater system after the last known case was identified. The decrease of RNA to undetectable levels was consistent with the previously reported median fecal shedding time of 25 days among hospitalized COVID-19 patients (9).
In total, 9 of 10 confirmed patients during the first outbreak and 10 of 15 patients during the second outbreak lived in residence halls where RNA was present in grab samples collected <7 days before diagnosis. Similarly, a study at a large college campus found the presence of RNA in grab samples from residence halls had 79.9% positive predictive value of COVID-19 cases within 4 days of collection (10). The RNA in 4 grab samples from halls D and F during days 45–53 could be from visitors recovering or actively infectious with SARS-CoV-2; this ambiguity is a limitation of wastewater surveillance. We did not evaluate the magnitude of the RNA concentrations in the grab samples because those samples represent RNA loading in the wastewater flow only at the time of sampling.
In conclusion, wastewater surveillance can indicate changes in SARS-CoV-2 transmission in the student population at a small residential college. Wastewater surveillance can quickly identify outbreaks, localize the detection of COVID-19 cases, and inform the management of resources for clinical surveillance testing.
Dr. Brooks is an assistant professor at Saint Joseph’s College of Maine, Standish, Maine, USA. Her research interests include the sources and persistence of fecal pollution in drinking water, recreational waters, and wastewater, as well as its potential effects on human health.
Acknowledgments
We thank Eilidh Sidaway and Brianna Shelley for laboratory support.
The Saint Joseph’s College of Maine Pandemic Response team financially supported this project.
References
- Ahmed W, Angel N, Edson J, Bibby K, Bivins A, O’Brien JW, et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci Total Environ. 2020;728:
138764 . DOIPubMedGoogle Scholar - Weidhaas J, Aanderud ZT, Roper DK, VanDerslice J, Gaddis EB, Ostermiller J, et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci Total Environ. 2021;775:
145790 . DOIPubMedGoogle Scholar - Gibas C, Lambirth K, Mittal N, Juel MAI, Barua VB, Roppolo Brazell L, et al. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci Total Environ. 2021;782:
146749 . DOIPubMedGoogle Scholar - Scott LC, Aubee A, Babahaji L, Vigil K, Tims S, Aw TG. Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ Res. 2021;200:
111374 . DOIPubMedGoogle Scholar - Centers for Disease Control and Prevention. Wastewater surveillance data reporting and analytics. 2021 [cited 2020 Sep 5]. https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/data-reporting-analytics.html
- Centers for Disease Control and Prevention. Ending home isolation for persons with COVID-19 not in healthcare settings. 2021 [cited 2020 Sep 20]. https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-in-home-patients.html
- Centers for Disease Control and Prevention. If you are sick or caring for someone. 2020 [cited 2020 Sep 20]. https://www.cdc.gov/coronavirus/2019-ncov/if-you-are-sick/index.html
- Wölfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Müller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–9. DOIPubMedGoogle Scholar
- Wang X, Zheng J, Guo L, Yao H, Wang L, Xia X, et al. Fecal viral shedding in COVID-19 patients: Clinical significance, viral load dynamics and survival analysis. Virus Res. 2020;289:
198147 . DOIPubMedGoogle Scholar - Betancourt WQ, Schmitz BW, Innes GK, Prasek SM, Pogreba Brown KM, Stark ER, et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci Total Environ. 2021;779:
146408 . DOIPubMedGoogle Scholar
Figure
Cite This ArticleOriginal Publication Date: August 31, 2021
1Current affiliation: St. Catherine University, St. Paul, Minnesota, USA.
Table of Contents – Volume 27, Number 12—December 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Yolanda M. Brooks, Saint Joseph’s College of Maine, 278 Whites Bridge Rd, Standish, ME 04084, USA
Top