Volume 27, Number 4—April 2021
Research
Genomic Surveillance of a Globally Circulating Distinct Group W Clonal Complex 11 Meningococcal Variant, New Zealand, 2013–2018
Figure 5
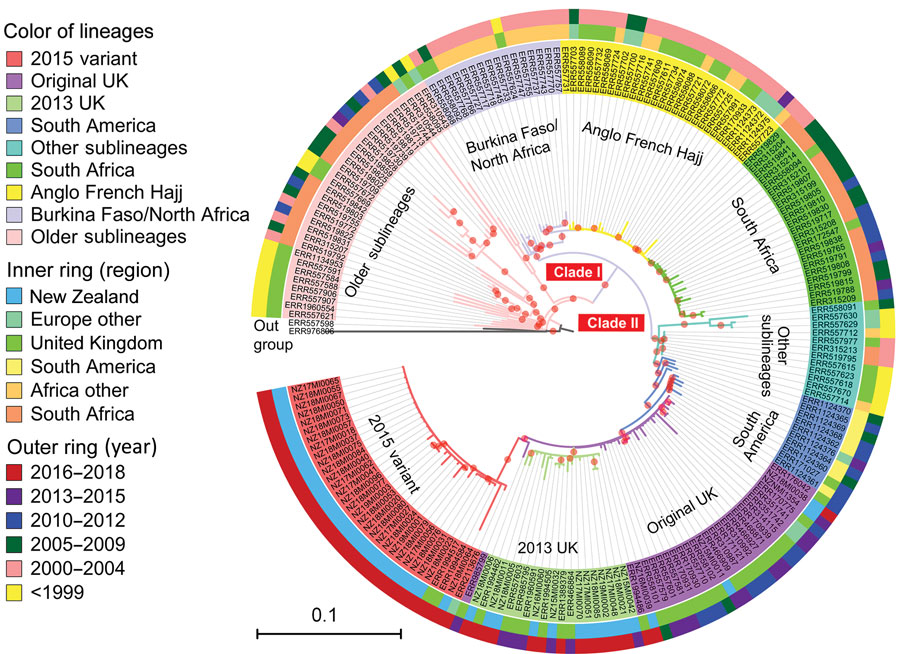
Figure 5. Phylogenetic position of New Zealand group W clonal complex 11 (W:CC11) Neisseria meningitidis isolates within the global W:CC11 major lineages. Maximum-likelihood phylogeny was generated by RAxML version 8.2.12 (33) on the basis of the core single-nucleotide polymorphism alignment of 198 W:CC11 isolates. Branches with a bootstrap (200 replications) value >90% are indicated with a red dot. Excluding the basal older sublineages, all other isolates form 2 strongly supported clades marked as clade I and clade II, which correspond to the Hajj strain sublineage and the South America strain sublineage. All the major defined lineages of W:CC11 are marked and indicated by consistent background color of isolate’s identification number and branches. The inner ring and outer ring designate the region and year of isolation for each isolate. Scale bar indicates average number of substitutions per site.
References
- Halperin SA, Bettinger JA, Greenwood B, Harrison LH, Jelfs J, Ladhani SN, et al. The changing and dynamic epidemiology of meningococcal disease. Vaccine. 2012;30(Suppl 2):B26–36. DOIPubMedGoogle Scholar
- Harrison LH, Trotter CL, Ramsay ME. Global epidemiology of meningococcal disease. Vaccine. 2009;27(Suppl 2):B51–63. DOIPubMedGoogle Scholar
- Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5. DOIPubMedGoogle Scholar
- Waśko I, Hryniewicz W, Skoczyńska A. Significance of meningococcal hyperinvasive clonal complexes and their influence on vaccines development. Pol J Microbiol. 2015;64:313–21. DOIPubMedGoogle Scholar
- Mustapha MM, Marsh JW, Krauland MG, Fernandez JO, de Lemos APS, Dunning Hotopp JC, et al. Genomic epidemiology of hypervirulent serogroup W, ST-11 Neisseria meningitidis. EBioMedicine. 2015;2:1447–55. DOIPubMedGoogle Scholar
- Ladhani SN, Beebeejaun K, Lucidarme J, Campbell H, Gray S, Kaczmarski E, et al. Increase in endemic Neisseria meningitidis capsular group W sequence type 11 complex associated with severe invasive disease in England and Wales. Clin Infect Dis. 2015;60:578–85. DOIPubMedGoogle Scholar
- Eriksson L, Hedberg ST, Jacobsson S, Fredlund H, Mölling P, Stenmark B. Whole-genome sequencing of emerging invasive Neisseria meningitidis serogroup W in Sweden. J Clin Microbiol. 2018;56:e01409–17. DOIPubMedGoogle Scholar
- Martin NV, Ong KS, Howden BP, Lahra MM, Lambert SB, Beard FH, et al.; Communicable Diseases Network Australia MenW Working Group. Rise in invasive serogroup W meningococcal disease in Australia 2013-2015. Commun Dis Intell Q Rep. 2016;40:E454–9.PubMedGoogle Scholar
- Tsang RSW, Ahmad T, Tyler S, Lefebvre B, Deeks SL, Gilca R, et al. Whole genome typing of the recently emerged Canadian serogroup W Neisseria meningitidis sequence type 11 clonal complex isolates associated with invasive meningococcal disease. Int J Infect Dis. 2018;69:55–62. DOIPubMedGoogle Scholar
- Potts CC, Joseph SJ, Chang HY, Chen A, Vuong J, Hu F, et al. Population structure of invasive Neisseria meningitidis in the United States, 2011-15. J Infect. 2018;77:427–34. DOIPubMedGoogle Scholar
- Lucidarme J, Hill DMC, Bratcher HB, Gray SJ, du Plessis M, Tsang RSW, et al. Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J Infect. 2015;71:544–52. DOIPubMedGoogle Scholar
- Taha MK, Achtman M, Alonso JM, Greenwood B, Ramsay M, Fox A, et al. Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet. 2000;356:2159. DOIPubMedGoogle Scholar
- Weidlich L, Baethgen LF, Mayer LW, Moraes C, Klein CC, Nunes LS, et al. High prevalence of Neisseria meningitidis hypervirulent lineages and emergence of W135:P1.5,2:ST-11 clone in Southern Brazil. J Infect. 2008;57:324–31. DOIPubMedGoogle Scholar
- Lucidarme J, Scott KJ, Ure R, Smith A, Lindsay D, Stenmark B, et al. An international invasive meningococcal disease outbreak due to a novel and rapidly expanding serogroup W strain, Scotland and Sweden, July to August 2015. Euro Surveill. 2016;21:15–23. DOIPubMedGoogle Scholar
- Krone M, Gray S, Abad R, Skoczyńska A, Stefanelli P, van der Ende A, et al. Increase of invasive meningococcal serogroup W disease in Europe, 2013 to 2017. Euro Surveill. 2019;24:24. DOIPubMedGoogle Scholar
- Devoy AF, Dyet KH, Martin DR. Stability of PorA during a meningococcal disease epidemic. J Clin Microbiol. 2005;43:832–7. DOIPubMedGoogle Scholar
- Dyet KH, Martin DR. Clonal analysis of the serogroup B meningococci causing New Zealand’s epidemic. Epidemiol Infect. 2006;134:377–83. DOIPubMedGoogle Scholar
- Sexton K, Lennon D, Oster P, Crengle S, Martin D, Mulholland K, et al. The New Zealand Meningococcal Vaccine Strategy: a tailor-made vaccine to combat a devastating epidemic. N Z Med J. 2004;117:U1015.PubMedGoogle Scholar
- Loring BJ, Turner N, Petousis-Harris H. MeNZB vaccine and epidemic control: when do you stop vaccinating? Vaccine. 2008;26:5899–904. DOIPubMedGoogle Scholar
- Institute of Environmental Science and Research Limited. Notifiable Diseases in New Zealand: Annual Report 2016. Porirua, New Zealand [cited 2020 Nov 3]. https://surv.esr.cri.nz/surveillance/annual_surveillance.php?we_objectID=4656
- Bennett DE, Cafferkey MT. Consecutive use of two multiplex PCR-based assays for simultaneous identification and determination of capsular status of nine common Neisseria meningitidis serogroups associated with invasive disease. J Clin Microbiol. 2006;44:1127–31. DOIPubMedGoogle Scholar
- Oksanen G, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: community ecology package [cited 2020 Nov 3]. https://CRAN.R-project.org/package=vegan
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Kwong JC, Gonçalves da Silva A, Stinear TP, Howden BP, Seemann T. Meningotype: [cited 2020 Nov 3]. https://github.com/MDU-PHL/meningotype
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. DOIPubMedGoogle Scholar
- Okonechnikov K, Conesa A, García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–4.PubMedGoogle Scholar
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al.; 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. DOIPubMedGoogle Scholar
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. DOIPubMedGoogle Scholar
- Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. DOIPubMedGoogle Scholar
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. DOIPubMedGoogle Scholar
- Felsenstein J. Confidence-limits on phylogenies—an approach using the bootstrap. Evolution. 1985;39:783–91. DOIPubMedGoogle Scholar
- Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLOS Comput Biol. 2015;11:
e1004041 . DOIPubMedGoogle Scholar - Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):
W242-5 . DOIPubMedGoogle Scholar - Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. DOIPubMedGoogle Scholar
- Bratcher HB, Corton C, Jolley KA, Parkhill J, Maiden MC. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics. 2014;15:1138. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 28th edition (informational supplement M100). Wayne (PA): The Institute; 2018.
- Zeileis A, Leisch F, Hornik K, Kleiber C, Hansen B, Zeileis MA. Package ‘strucchange’ [cited 2020 Nov 3]. https://CRAN.R-project.org/package=strucchange
- Mowlaboccus S, Jolley KA, Bray JE, Pang S, Lee YT, Bew JD, et al. Clonal expansion of new penicillin-resistant clade of Neisseria meningitidis serogroup W clonal complex 11, Australia. Emerg Infect Dis. 2017;23:1364–7. DOIPubMedGoogle Scholar
- Whaley MJ, Joseph SJ, Retchless AC, Kretz CB, Blain A, Hu F, et al. Whole genome sequencing for investigations of meningococcal outbreaks in the United States: a retrospective analysis. Sci Rep. 2018;8:15803. DOIPubMedGoogle Scholar
- Public Health England. Invasive meningococcal disease in England: annual laboratory confirmed reports for epidemiological year 2017 to 2018. London: Public Health England; 2018 [cited 2020 Nov 3]. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/751821/hpr3818_IMD.pdf
- Knol MJ, de Melker HE, Berbers GAM, van Ravenhorst MB, Ruijs WLM, van Vliet JA. Meningococcal disease in the Netherlands: background information for the Health Council. RIVM Report 2017–0031. 2017 [cited 2020 Nov 03]. https://www.rivm.nl/bibliotheek/rapporten/2017-0031.pdf
- Knol MJ, Hahné SJM, Lucidarme J, Campbell H, de Melker HE, Gray SJ, et al. Temporal associations between national outbreaks of meningococcal serogroup W and C disease in the Netherlands and England: an observational cohort study. Lancet Public Health. 2017;2:e473–82. DOIPubMedGoogle Scholar
- Leimkugel J, Racloz V, Jacintho da Silva L, Pluschke G. Global review of meningococcal disease. A shifting etiology. J Bacterial Res. 2009;1:6–18.
- Bröker M, Jacobsson S, Kuusi M, Pace D, Simões MJ, Skoczynska A, et al. Meningococcal serogroup Y emergence in Europe: update 2011. Hum Vaccin Immunother. 2012;8:1907–11. DOIPubMedGoogle Scholar
- Ministry of Health Manatū Hauora. Change to treatment recommendations for meningococcal disease. 2018 Nov 30 [cited 2020 Nov 3]. https://www.health.govt.nz/news-media/news-items/change-treatment-recommendations-meningococcal-disease
1These authors contributed equally to this article.