Volume 28, Number 10—October 2022
Dispatch
Endofungal Mycetohabitans rhizoxinica Bacteremia Associated with Rhizopus microsporus Respiratory Tract Infection
Cite This Article
Citation for Media
Abstract
We report Mycetohabitans rhizoxinica bacteremia in a 65-year-old woman in California, USA, who was undergoing chimeric antigen receptor T-cell therapy for multiple myeloma. Acute brain infarction and pneumonia developed; Rhizopus microsporus mold was isolated from tracheal suction. Whole-genome sequencing confirmed bacteria in blood as genetically identical to endofungal bacteria inside the mold.
Mycetohabitans rhizoxinica (previously Burkholderia rhizoxinica) and M. endofungorum are endofungal bacteria inhabiting Rhizopus microsporus mold. These bacteria form symbiosis to help the mold infect plants and produce mycotoxins, such as rhizoxin, that cause rice seedling blight (1,2). Genetically, Mycetohabitans species are highly similar to Burkholderia species but with significantly smaller genome sizes (3.3–3.8 Mb vs. 5.8–11 Mb), reflecting their endosymbiotic nature (3). R. microsporus causes mucormycosis, a devastating invasive fungal infection seen most prevalently in immunocompromised patients, but no strong evidence has suggested that the symbiont bacteria contribute to the pathogenicity of R. microsporus mold in humans (1,4). Isolation of endofungal bacteria have seldom been reported in clinical settings, most likely because of limitations in identification methods. In a study performed by the Centers for Disease Control and Prevention, M. rhizoxinica and M. endofungorum isolated from blood and wounds were characterized as oxidase-positive, gram-negative coccobacilli and could be reliably identified only by sequencing; no clinical information was provided to characterize the clinical presentation nor did researchers describe any link to R. microsporus (4). We report M. rhizoxinica bacteremia associated with multifocal pneumonia presumptively caused by R. microsporus mold in a severely immunocompromised patient.
A 65-year-old woman in California, USA, with a history of relapsed and refractory multiple myeloma visited a hospital to receive chimeric antigen receptor (CAR) T-cell therapy. She previously had received 3 doses of the COVID-19 mRNA vaccine. On the day after CAR T-cell therapy, she experienced a rapid decline in mental status, accompanied by fever and hypotension, and was transferred to the intensive care unit. At that time, her total leukocyte count was 0.09 × 103 cells/uL. Nasopharyngeal testing for COVID-19 by PCR was negative. She was diagnosed with immune effector cell-associated neurotoxicity syndrome and cytokine release syndrome. She received high doses of dexamethasone and 4 doses of tocilizumab, as well as anakinra, which was tapered over 7 days. The patient’s symptoms improved, and she was discharged after a 15-day hospital stay, with plans for a prolonged taper of orally administered dexamethasone (8 mg 2×/d). On discharge, the patient’s leukocyte count was 1.4 × 103 cells/uL (87% neutrophils, 7% lymphocytes); the next day, her outpatient blood work showed mild lactic acidosis.
Four days after leaving the hospital, the patient sought treatment at an outpatient oncology clinic, reporting generalized weakness and fatigue. We performed blood and urine analyses and began a regimen of oral levofloxacin (500 mg daily). On day 6 after discharge, we noted a positive blood culture result, with a gram-negative rod. The urine culture grew 30,000 CFU/mL of Klebsiella pneumoniae. When evaluating the patient during clinical rounds, we performed repeat blood and urine cultures and recommended inpatient management, which was refused. We administered 1 intravenous dose of meropenem (1 g) at that visit and 1 dose of intravenous ceftriaxone (2 g) the next day.
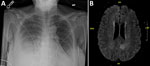
On day 8 after discharge, the patient sought emergency treatment for worsening foot and ankle pain. Her leukocyte count was 0.44 × 103 cells/uL. Bacterial identification of the positive blood culture was still pending. Results of repeat blood and urine cultures and COVID-19 PCR testing (nasopharyngeal) were negative; chest radiograph showed right-, middle-, and lower-lobe airspace opacification (Figure 1, panel A). We began piperacillin/tazobactam and performed an arthrocentesis, which showed crystals but revealed a negative Gram stain. Shortly thereafter, the patient reported acute numbness and weakness of her right leg. She subsequently developed dyspnea, respiratory distress, and altered mental status. We intubated her and conducted magnetic resonance imaging, which showed an acute infarct of the left medial parietal lobe, with hemorrhagic transformation (Figure 1, panel B). We transferred the patient to the intensive care unit, where she was febrile (38.2°C) and required 2 vasopressors. We administered vancomycin, caspofungin, and isavuconazole. The next day, hypotension worsened, requiring 3 vasopressors. Leukocyte was 0.16 × 103 cells/uL. On hospital day 3, we obtained a tracheal aspirate for cultural analysis. Later that day, pulseless electrical activity occurred; the patient suffered cardiac arrest and died.
The day after her death, results of analysis of tracheal aspirate obtained on hospital day 3 revealed a mold. The bacteria from the initial positive blood culture were gram-negative, oxidase-positive coccobacilli not identifiable by the Vitek MS system (bioMérieux, https://www.biomerieux.com). The specimen was automatically reflexed to a laboratory-developed whole-genome sequencing (WGS) identification test (5). Drug-susceptibility testing showed presumptive (due to lack of breakpoint) susceptibility to most drugs tested, including amoxicillin/clavulanate, ceftriaxone, piperacillin/tazobactam, carbapenems, gentamicin, ciprofloxacin, and trimethoprim/sulfamethoxazole (Table).
We performed WGS on the bacteria from both the blood and the mold obtained from tracheal suction using Illumina MiSeq (Illumina, https://www.illumina.com) (5) (Figure 2). We identified the bacteria as M. rhizoxinica, with >99% identity in all 3 full-length marker genes, including 16S, rpoB, and groL (hsp65) compared with the type strain M. rhizoxinica HKI 454 (GenBank accession no. NC_014722.1). We identified the mold as R. microsporus, with >99% identity in the internal transcribed spacer (ITS) gene and the D1–D2 region of the 28S gene compared with R. microsporus var. chinensis CBS 631.82 (accession no. NR_149337.1 for ITS and HM849668.1 for D1–D2). We mapped the whole genome sequences of the bacteria and mold isolates (Genbank Sequence Read Archive data: PRJNA857096) to the M. rhizoxinica HKI 454 complete genome using Geneious Prime (Geneious, https://www.geneious.com) and achieved similar whole-genome coverage (bacteria, 94.0%; mold, 94.1%) and pairwise identity (bacteria, 95.5%; mold, 95.8%), with sufficient mean coverage (bacteria, 298×; mold, 75×). We used the mapped sequence reads from the bacteria and the mold for single-nucleotide polymorphism analyses using CLC Genomics Workbench (QIAGEN, https://www.qiagen.com) as previously described (6). Results showed no single-nucleotide polymorphism between the sequences in the bacteria from blood and the bacteria within the mold, indicating the bacteria in the blood was derived from the mold (Figure 2). Further genomic analysis confirmed the presence of a gene cluster (rhiA–rhiI) that encodes the biosynthesis of rhizoxin in both bacterial and mold isolates (7).
Using WGS, we present clear evidence linking the endofungal bacteria M. rhizoxinica, isolated from a patient’s blood, to the bacteria residing within the R. microsporus mold isolated from the patient’s respiratory sample. The bacteria were isolated in the blood culture before initiating antibiotics were pansusceptible to the antibiotics tested, and cleared quickly after treatment. However, it is not clear whether the antibiotics retained activity against the bacteria within the fungal cytoplasm or whether activity was preserved for only bacteria outside the fungi. We hypothesize that the bacteria were most likely inside the mold in vivo and freed only after sample collection and then grew in the blood culture bottle during the incubation, when the fungus became degraded or lysed. Blood culture has low yield for Mucorales species (8), indicating those molds often die during the blood culture process. Because of the clinical manifestation, we believe the bacteria did not contribute to sepsis but rather served as a signal for a developing invasive mold infection that manifested during the immune suppression related to the patient’s CAR T-cell therapy. Unfortunately, the patient decompensated rapidly and died before the mold was identified and before we could initiate aggressive antifungal treatment.
Infarction and necrosis of infected tissues are hallmarks of mucormycosis (9). Disseminated mucormycosis, the presumptive diagnosis in this case based on the pulmonary findings and hemorrhagic brain infarction, is a rapidly progressive infection associated with a mortality rate >90% (10). This patient was at risk for disseminated mucormycosis because of profound immune suppression. Early diagnosis and prompt treatment of mucormycosis are key to improving clinical outcomes. Disseminated mucormycosis appears to be the underlying infectious process in this case, but this report lacks autopsy examination to confirm the presumptive diagnosis.
In conclusion, we found that isolation of endofungal bacteria M. rhizoxinica in the clinical setting might indicate invasive Rhizopus infection. Because identification methods used in most clinical laboratories are limited, endofungal bacteria may be underrecognized and require sequencing to identify.
Dr. Yang is a clinical microbiologist and assistant clinical professor at the University of California–Los Angeles Department of Pathology and Medicine. Dr. Yang's main research interests include molecular diagnostics and innovation, clinical virology, transplant-related infectious diseases testing, genomic epidemiology, and antimicrobial resistance mechanisms.
Acknowledgments
We thank the University of California–Los Angeles Clinical Microbiology Laboratory for technical support.
This work was supported by the National Institutes of Health (K01TW012170 to PCA).
References
- Frey-Klett P, Burlinson P, Deveau A, Barret M, Tarkka M, Sarniguet A. Bacterial-fungal interactions: hyphens between agricultural, clinical, environmental, and food microbiologists. Microbiol Mol Biol Rev. 2011;75:583–609. DOIPubMedGoogle Scholar
- Lackner G, Hertweck C. Impact of endofungal bacteria on infection biology, food safety, and drug development. PLoS Pathog. 2011;7:
e1002096 . DOIPubMedGoogle Scholar - Estrada-de Los Santos P, Palmer M, Chávez-Ramírez B, Beukes C, Steenkamp ET, Briscoe L, et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes (Basel). 2018;9:389. DOIPubMedGoogle Scholar
- Gee JE, Glass MB, Lackner G, Helsel LO, Daneshvar M, Hollis DG, et al. Characterization of Burkholderia rhizoxinica and B. endofungorum isolated from clinical specimens. PLoS One. 2011;6:
e15731 . DOIPubMedGoogle Scholar - Price TK, Realegeno S, Mirasol R, Tsan A, Chandrasekaran S, Garner OB, et al. Validation, implementation, and clinical utility of whole genome sequence-based bacterial identification in the clinical microbiology laboratory. J Mol Diagn. 2021;23:1468–77. DOIPubMedGoogle Scholar
- Price TK, Mirasol R, Ward KW, Dayo AJ, Hilt EE, Chandrasekaran S, et al. Genomic characterizations of clade iii lineage of Candida auris, California, USA. Emerg Infect Dis. 2021;27:1223–7. DOIPubMedGoogle Scholar
- Partida-Martinez LP, Hertweck C. A gene cluster encoding rhizoxin biosynthesis in “Burkholderia rhizoxina”, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem. 2007;8:41–5. DOIPubMedGoogle Scholar
- Walsh TJ, Gamaletsou MN, McGinnis MR, Hayden RT, Kontoyiannis DP. Early clinical and laboratory diagnosis of invasive pulmonary, extrapulmonary, and disseminated mucormycosis (zygomycosis). Clin Infect Dis. 2012;54(Suppl 1):S55–60. DOIPubMedGoogle Scholar
- Petrikkos G, Skiada A, Lortholary O, Roilides E, Walsh TJ, Kontoyiannis DP. Epidemiology and clinical manifestations of mucormycosis. Clin Infect Dis. 2012;54(Suppl 1):S23–34. DOIPubMedGoogle Scholar
- Roden MM, Zaoutis TE, Buchanan WL, Knudsen TA, Sarkisova TA, Schaufele RL, et al. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin Infect Dis. 2005;41:634–53. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: September 12, 2022
Table of Contents – Volume 28, Number 10—October 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Address for correspondence: Shangxin Yang, UCLA Clinical Microbiology Laboratory, 11633 San Vicente Blvd, Los Angeles, CA 90049, USA
Top