Volume 28, Number 11—November 2022
Dispatch
Molecular Detection of Haplorchis pumilio Eggs in Schoolchildren, Kome Island, Lake Victoria, Tanzania
Figure 2
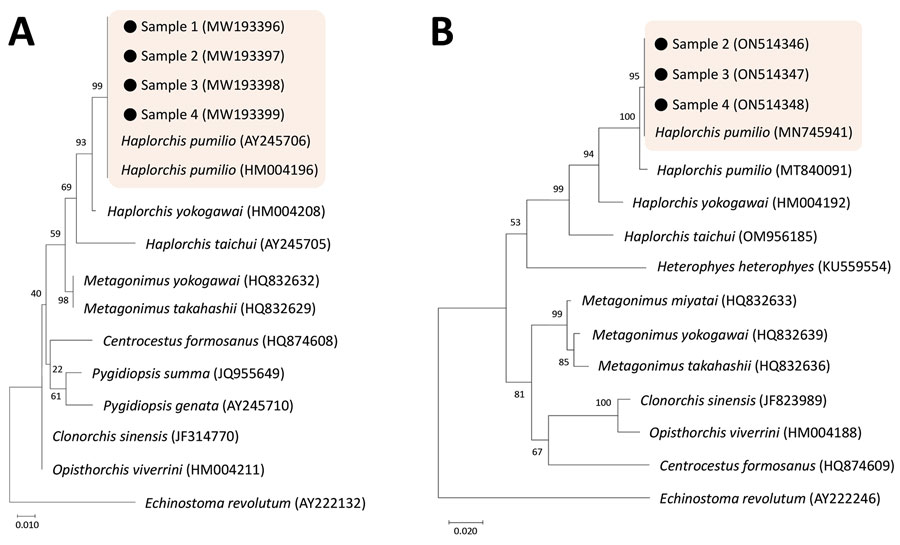
Figure 2. Phylogenetic trees of DNA of small trematode eggs from schoolchildren, Kome Island, Lake Victoria, Tanzania, in comparison with reference sequences of heterophyid (Haplorchis pumilio and others) and opisthorchiid trematodes, based on 18S (A) and 28S rDNA (B) sequences. The trees were constructed using the maximum-likelihood method based on the Kimura 2-parameter model and viewed by the MEGA 7.0 program (http://www.megasoftware.net). GenBank accession numbers are indicated. Scale bars indicate nucleotide substitutions per site.
1These authors contributed equally to this article.
Page created: September 07, 2022
Page updated: October 24, 2022
Page reviewed: October 24, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.