Volume 28, Number 2—February 2022
Dispatch
SARS-CoV-2 Cross-Reactivity in Prepandemic Serum from Rural Malaria-Infected Persons, Cambodia
Abstract
Inhabitants of the Greater Mekong Subregion in Cambodia are exposed to pathogens that might influence serologic cross-reactivity with severe acute respiratory syndrome coronavirus 2. A prepandemic serosurvey of 528 malaria-infected persons demonstrated higher-than-expected positivity of nonneutralizing IgG to spike and receptor-binding domain antigens. These findings could affect interpretation of large-scale serosurveys.
Serosurveys for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the Greater Mekong Subregion (GMS) of Cambodia have been limited to those screening healthcare workers in 2 urban hospital-based settings (1,2). These antibody-based studies are necessary to determine at-risk populations and direct disease containment measures; however, before informing public health decisions, serologic assays require careful, country-specific calibration because several regions report fluctuating results or high background reactivity in different populations (3–5). This variability might be attributable to myriad serologic assays, the hypothesized cross-reactivity from common cold–type respiratory coronaviruses (6), previous Plasmodium infections (7,8; S. Lapidus et al., unpub. data, https://www.medrxiv.org/content/10.1101/2021.05.10.21256855v1), or previously uncharacterized betacoronaviruses in wildlife populations in the rural GMS (9–11). Although many serologic SARS-CoV-2 investigations are in progress, considering how pathogen diversity in the GMS might influence estimations of SARS-CoV-2 seroprevalence is prudent.
We tested serum or plasma samples collected from 528 malaria-infected persons in Cambodia during 2005–2011 (before SARS-CoV-2 emerged in 2019) for IgG reactive to SARS-CoV-2 spike and receptor-binding domain (RBD) proteins by using ELISA (12,13). We used de-identified, anonymized serum or plasma samples biobanked after malaria research studies (NCT00341003, NCT00663546, and NCT01350856, approved by the National Institute of Allergy and Infectious Diseases and the National Ethics Committee on Human Research in Cambodia) for this retrospective study.
Because 6 other coronaviruses (OC43, HKU1, 229E, NL63, severe acute respiratory syndrome coronavirus 1 [SARS-CoV-1], and Middle East respiratory syndrome coronavirus) possess structural proteins capable of infecting humans, we selected highly specific ELISAs for the SARS-CoV-2 structural proteins (12,13). Compared with other coronaviruses, SARS-CoV-2 shows varying levels of spike protein sequence homology; levels are highest for SARS-CoV-1 (76% identity, 87% similarity) and lowest for the common cold coronavirus HKU1 (29% identity, 40% similarity) (12). Reactivity to both spike and RBD antigens above cutoff values is required for a positive test with reported sensitivity of 100% (95% CI 92.9%–100%) and specificity of 100% (95% CI 98.8%–100%) (12,13). Prepandemic samples had levels above the set cutoffs for SARS-CoV-2 spike and RBD antigens (Figure 1) varying from 4.4% to 13.8% positivity to both SARS-CoV-2 spike and RBD depending on which cutoff values (calibrated for the Mali or US populations) were used for this assay (4,12,13) (Table; Figure 1; Appendix Table 1).
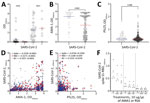
To test whether the higher-than-expected positivity was an artifact of our in-house ELISA, we tested a subset of samples with a commercially validated SARS-CoV-2 Spike S1-RBD IgG ELISA Detection Kit (Genscript, https://www.genscript.com). Of the 24 persons who were seronegative by in-house assay and 11 who were seropositive by in-house assay, 18 tested negative and 9 tested positive by the commercial test, yielding an overall concordance of 77.1% between assays (Appendix Table 2). This inconsistency might be explained by the stringency of the in-house assay that tests both spike and RBD versus the commercial kit that tests for RBD only; nevertheless, higher-than-expected positivity was observed in both assays. Since common cold coronaviruses do circulate in Cambodia, but no cases of SARS-CoV-1 or Middle East respiratory syndrome have been documented, we tested a subset of the cohort for IgG to HKU1 and OC43. Reactivity between subjects was comparable despite SARS-CoV-2 serostatus (Figure 2, panel A).
We further tested 289 samples to assess whether a relationship existed between antibodies to Plasmodium spp. and SARS-CoV-2 proteins by using 2 known malarial antigens: Plasmodium falciparum apical membrane antigen 1 (AMA-1), which is highly immunogenic and an indicator of parasite exposure, and P. falciparum Pfs25 protein (Pfs25), which is poorly immunogenic and expressed only during the mosquito stages of parasite development (4) (Figure 2, panels B–E). Of note, when we grouped samples by SARS-CoV-2 serostatus, we detected significantly higher levels of AMA-1 antibodies in SARS-CoV-2–seropositive persons than seronegative persons (mean AMA-1 antibody level 3.0 vs. 2.1; p = 0.0003) (Figure 2, panel B). As expected, no difference was seen in antibody levels to Pfs25 with regard to SARS-CoV-2 seropositivity (Figure 2, panel C). A weak but statistically significant positive correlation was detected between spike and RBD with AMA-1 IgG (Figure 2, panel D). This finding corroborates recent observations that higher SARS-CoV-2 seroreactivity by ELISA or rapid tests is detected in persons from malaria-endemic areas, expanding previous observations to include Southeast Asia (7,8; S. Lapidus et al., unpub. data). We also evaluated samples for seroreactivity against the nucleocapsid protein that also positively correlated with the AMA-1 IgG. Only nucleocapsid antibodies were weakly correlated with Pfs25 antibodies, which reinforces the argument for nonspecific nucleocapsid reactivity (Figure 2, panel E). Preincubation with 10 mg/mL of AMA-1 or bovine serum albumin had no notable effect on reactivity to SARS-CoV-2 spike S1-RBD (Figure 2, panel F). Therefore, Plasmodium spp. exposure might contribute to SARS-CoV-2 malaria-related background reactivity. This reactivity could be attributed to immune responses to other Plasmodium spp. proteins, polyclonal B cell activation during infection, or interaction with the sialic acid moiety on N-linked glycans of the SARS-CoV-2 spike protein (7; S. Lapidus et al., unpub. data). Of note, SARS-CoV-2 spike proteins used in the assays were produced in HEK293 mammalian cells and likely have comparable glycosylation patterns. Elsewhere, malaria-induced cross-reactivity in prepandemic samples from malaria-experienced persons from Africa was mitigated by the modification of 2 commercial assays to add a urea wash (S. Lapidus et al., unpub. data).
To elucidate the functionality of the detected antibodies, we took a subset (n = 21) of the samples with the highest reactivity to SARS-CoV-2 total IgG and performed neutralization assays (Appendix Figure). No neutralizing activity was identified despite high levels of antibodies reacting to both spike and RBD proteins. Identical results were obtained by using a surrogate virus neutralization test targeting RBD interaction with the host cell receptor ACE2 (Genscript) (Appendix Table 3) (14). Both SARS-CoV-2 infection and vaccination can trigger high levels of nonneutralizing antibodies, whereas neutralizing antibodies aimed primarily at the RBD seem to wane faster and remain at low titers (14). Plausibly, the cross-reactive nonfunctional antibodies to SARS-CoV-2 were raised during an infection by Plasmodium spp. (S. Lapidus et al., unpub. data), but we cannot discard the hypothesis that nonneutralizing SARS-CoV-2–reactive antibodies in prepandemic serum samples might be linked to the ability of betacoronaviruses to evade immune recognition because of their complex surfaces (14,15). A limitation in understanding the assays’ specificity is the lack of prepandemic samples from non–malaria-endemic areas and from present-day confirmed SARS-CoV-2 convalescent samples in Cambodia.
We found in a widely used, highly specific, and validated ELISA that ≈4%–14% of prepandemic serum samples from malaria-infected persons in Cambodia were positive for nonneutralizing antibodies to SARS-CoV-2 spike and RBD antigens by using various standardized optical density cutoff values (4,12,13). We noted a relationship between increased SARS-CoV-2 seroreactivity and antimalarial humoral immunity, which was also recently shown in Africa (S. Lapidus et al., unpub. data). The plausibility of regular spillover events, or simply increased exposure to uncharacterized betacoronaviruses, as a reason for SARS-CoV-2 cross-reactivity is also increased in settings at high risk for zoonotic disease transmission because of agricultural and dietary practices such as bat guano collection and consumption of wild meats (9–11). Given that 50%–80% of GMS residents are classified as rural, careful calibration of serologic assays targeting SARS-CoV-2 will be necessary in national and subnational serosurveys. Although neutralization assays with live virus are often considered the standard because of their specificity, they are cost-prohibitive for large-scale serosurveys. The use of competition ELISA assays such as surrogate virus neutralization tests targeting the RBD-ACE2 blockade might be an attractive option for populations at high risk for zoonotic exposures in resource-scarce settings without Biosafety Level 3 facilities.
Dr. Manning is an infectious diseases physician-scientist focused on vector-borne disease epidemiology. She is currently based in Phnom Penh as an Assistant Clinical Investigator for the National Institute of Allergy and Infectious Diseases and its International Center of Excellence in Research Cambodia.
Acknowledgments
We thank the participants of the studies in Pursat, Ratanakiri, and Preah Vihear provinces and the original study protocol staff.
This research was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases.
References
- Song LH, Hoan NX, Bang MH, Kremsner PG, Velavan TP. Viral and serological testing of SARS-CoV-2 among health care workers and patients in Vietnam. Lancet Reg Health West Pac. 2021;8:
100113 . DOIPubMedGoogle Scholar - Nopsopon T, Pongpirul K, Chotirosniramit K, Jakaew W, Kaewwijit C, Kanchana S, et al. Seroprevalence of hospital staff in a province with zero COVID-19 cases. PLoS One. 2021;16:
e0238088 . DOIPubMedGoogle Scholar - Uyoga S, Adetifa IMO, Karanja HK, Nyagwange J, Tuju J, Wanjiku P, et al. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Kenyan blood donors. Science. 2021;371:79–82. DOIPubMedGoogle Scholar
- Sagara I, Woodford J, Dicko A, Zeguime A, Doucoure M, Kwan J, et al. Rapidly increasing SARS-CoV-2 seroprevalence and limited clinical disease in 3 Malian communities: a prospective cohort study. Clin Infect Dis. 2021; [Epub ahead of print]. DOIGoogle Scholar
- Murhekar MV, Bhatnagar T, Selvaraju S, Saravanakumar V, Thangaraj JWV, Shah N, et al.; ICMR Serosurveillance Group. SARS-CoV-2 antibody seroprevalence in India, August-September, 2020: findings from the second nationwide household serosurvey. Lancet Glob Health. 2021;9:e257–66. DOIPubMedGoogle Scholar
- Ng KW, Faulkner N, Cornish GH, Rosa A, Harvey R, Hussain S, et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–43. DOIPubMedGoogle Scholar
- Steinhardt LC, Ige F, Iriemenam NC, Greby SM, Hamada Y, Uwandu M, et al. Cross-reactivity of two SARS-CoV-2 serological assays in a setting where malaria is endemic. J Clin Microbiol. 2021;59:
e0051421 . DOIPubMedGoogle Scholar - Vanroye F, Bossche DVD, Brosius I, Tack B, Esbroeck MV, Jacobs J. COVID-19 antibody detecting rapid diagnostic tests show high cross-reactivity when challenged with pre-pandemic malaria, schistosomiasis and Dengue samples. Diagnostics (Basel). 2021;11:1163. DOIPubMedGoogle Scholar
- Delaune D, Hul V, Karlsson EA, Hassanin A, Ou TP, Baidaliuk A, et al. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat Commun. 2021;12:6563. DOIPubMedGoogle Scholar
- Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. [Erratum in: Nat Commun. 2021;12:1430]. Nat Commun. 2021;12:972. DOIPubMedGoogle Scholar
- Huong NQ, Nga NTT, Long NV, Luu BD, Latinne A, Pruvot M, et al. Coronavirus testing indicates transmission risk increases along wildlife supply chains for human consumption in Viet Nam, 2013-2014. PLoS One. 2020;15:
e0237129 . DOIPubMedGoogle Scholar - Klumpp-Thomas C, Kalish H, Drew M, Hunsberger S, Snead K, Fay MP, et al. Standardization of ELISA protocols for serosurveys of the SARS-CoV-2 pandemic using clinical and at-home blood sampling. Nat Commun. 2021;12:113. DOIPubMedGoogle Scholar
- Hicks J, Klumpp-Thomas C, Kalish H, Shunmugavel A, Mehalko J, Denson J-P, et al. Serologic cross-reactivity of SARS-CoV-2 with endemic and seasonal betacoronaviruses. J Clin Immunol. 2021;41:906–13. DOIPubMedGoogle Scholar
- Tan CW, Chia WN, Qin X, Liu P, Chen MI-C, Tiu C, et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol. 2020;38:1073–8. DOIPubMedGoogle Scholar
- Bachmann MF, Mohsen MO, Zha L, Vogel M, Speiser DE. SARS-CoV-2 structural features may explain limited neutralizing-antibody responses. NPJ Vaccines. 2021;6:2. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: January 13, 2022
Table of Contents – Volume 28, Number 2—February 2022
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Fabiano Oliveira, National Institute of Allergy and Infectious Diseases, 12735 Twinbrook Pkwy, Rockville, MD 20852, USA
Top