Volume 28, Number 7—July 2022
Research
Outbreak of IncX8 Plasmid–Mediated KPC-3–Producing Enterobacterales Infection, China
Abstract
Carbapenem-resistant Enterobacterales (CRE) infection is highly endemic in China; Klebsiella pneumoniae carbapenemase (KPC) 2–producing CRE is the most common, whereas KPC-3–producing CRE is rare. We report an outbreak of KPC-3–producing Enterobacterales infection in China. During August 2020–June 2021, 25 blaKPC-3–positive Enterobacteriale isolates were detected from 24 patients in China. Whole-genome sequencing analysis revealed that the blaKPC-3 genes were harbored by IncX8 plasmids. The outbreak involved clonal expansion of KPC-3–producing Serratia marcescens and transmission of blaKPC-3 plasmids across different species. The blaKPC-3 plasmids demonstrated high conjugation frequencies (10−3 to 10−4). A Galleria mellonella infection model showed that 2 sequence type 65 K2 K. pneumoniae strains containing blaKPC-3 plasmids were highly virulent. A ceftazidime/avibactam in vitro selection assay indicated that the KPC-3–producing strains can readily develop resistance. The spread of blaKPC-3–harboring IncX8 plasmids and these KPC-3 strains should be closely monitored in China and globally.
Carbapenemase-producing Enterobacterales (CPE) have emerged as important nosocomial pathogens and are a global public health concern because of the high prevalence of CRE infection and its associated mortality rate. Currently, Klebsiella pneumoniae carbapenemase (KPC) is the most clinically important carbapenemase globally (1,2). Since its first discovery in 1996 (3), more than 90 KPC variants have been documented, of which KPC-2 and KPC-3 are the most common clinical variants (4). blaKPC genes are frequently harbored by Tn4401 or non-Tn4401 mobile elements (NTMKPC) (5), and the spread of blaKPC has been primarily associated with transmissible plasmids, belonging to different incompatibility groups (e.g., IncFII, IncI2, IncX, IncA/C, IncR, IncN, and ColE) (5).
China is regarded as a CRE-endemic region where K. pneumoniae, Escherichia coli, and Enterobacter cloacae complex are the most common CRE species (6,7). Among the CREs, ≈80%–90% were carbapenemase-producers, including >90% carbapenem-resistant K. pneumoniae and E. coli and ≈80% of carbapenem-resistant E. cloacae strains. For carbapenemase genes, the blaKPC-2 was the most dominant type (≈60%), followed by blaNDMs and blaIMPs. Interestingly, other KPC variants, especially KPC-3, are rarely detected in China. Compared with KPC-2, KPC-3 differs by a single amino acid substitution (H272Y) and shows higher hydrolysis efficiency against oxyimino-cephalosporins and carbapenems (8). In most KPC-endemic regions, including United States and countries in Europe, KPC-3 showed similar prevalence as that of KPC-2 enzyme, and both KPC variants were frequently detected in clinical CRE isolates. Despite China being KPC-endemic, KPC-3 has only been sporadically reported in China (9–13).
In this study, we describe a hospital outbreak of KPC-3–producing Enterobacterales involved with multiple species, including Serratia marcescens, K. pneumoniae, Escherichia coli, Enterobacter hormaechei, and Proteus mirabilis in mainland China. We obtained approval for the study from Ningbo First Hospital Ethics Committee (approval no. 2021RS095).
During August 1, 2020–June 30, 2021, we collected from a tertiary hospital in Ningbo, Zhejiang Province, China, 25 nonrepeated KPC-3–producing Enterobacterales isolates showing reduced susceptibility to carbapenems. None of the patients from whom the isolates were taken had international travel history in the preceding 3 months. We detected the presence of carbapenemase genes, including blaKPC, blaNDM, blaOXA-48–like, blaVIM, and blaIMP, by using PCR, followed by Sanger sequencing (14,15). We initially determined speciation by using matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and analyzed results by using the Vitek MS database (bioMérieux, https://www.biomerieux.com); we later confirmed the results by using whole-genome sequencing (WGS) analysis.
We performed antimicrobial susceptibility testing and modified carbapenem inactivation and applied WGS to explore the molecular features of the isolates (Appendix 1). We also conducted conjugation and electroporation experiments (and performed pulsed-field gel electrophoresis (PFGE) and S1-nuclease PFGE (Appendix 1). We applied string test to examine the Hypermucoviscous phenotypes of K. pneumoniae strains and Galleria mellonella infection model to evaluate the virulence potential of sequence type (ST) 65 K2 K. pneumoniae strains (Appendix 1). We applied a ceftazidime/avibactam in vitro selection assay to evaluate whether the KPC-3-producing strains were easily selected to be resistant to ceftazidime/avibactam (Appendix 1).
We submitted the complete nucleotide sequences of the plasmids pFK3112-KPC-3 and pCG2111-KPC-3 to GenBank (accession nos. CP081509 [pFK3112-KPC-3] and CP081510 [pCG2111-KPC-3]). We also deposited the raw reads of the genomes we sequenced in GenBank (Bioproject accession no. PRJNA354234).
Outbreak Description of blaKPC-3–harboring Enterobacterales
During August 1, 2020–June 30, 2021, we detected 25 KPC-3–producing Enterobacterales isolates in patients of a tertiary hospital in eastern China, including 18 Serratia marcescens, 3 K. pneumoniae, 1 E. coli, 2 E. hormaechei, and 1 Proteus mirabilis. The 25 isolates were from 24 patients; 1 patient had 2 isolates from sputum (FK3015) and blood (FK3018). The strains were recovered from sputum (n = 19), blood (n = 3), urine (n = 1), puncture fluid (n = 1), and bile (n = 1).
The first KPC-3 strain (S. marcescens CG2008) was isolated in August 2020, and the patient was admitted to the intensive care unit (ICU) 1 (building 3) with unconsciousness attributable to a head injury sustained in an accident. After 20 days of the patient’s hospitalization, we detected a carbapenem-resistant S. marcescens in the patient’s sputum. After that, we detected 4 additional carbapenem-resistant S. marcescens strains in the same ICU ward during August 2020–May 2021 (Appendix 2 Table 1). Starting in January 2021, we also found carbapenem-resistant S. marcescens strains (n = 6) in another ICU (building 2, ICU-2) and the wards of cardiology (building 2) (n = 2), emergency (building 3) (n = 1), and hepatobiliary and pancreatic surgery (building 3) (n = 1). In addition, starting in September 2020, we identified these KPC-3–producing strains in other Enterobacterales species in ICU-2 (K. pneumoniae), the coronary heart disease care unit (K. pneumoniae), ICU-1 (P. mirabilis and E. coli), the infectious disease ward (building 6) (E. hormaechei), and the hepatobiliary and pancreatic surgery ward (E. hormaechei) (Appendix 2 Table 1).
Of the patients, 23/24 were admitted into wards in medical buildings 2 and 3, including the 2 ICUs, in our hospital (Figure 1, panel A); 18 of them were infected with the S. marcescens (blaKPC-3). Most patients shared the same ward during the same time, especially the patients from ICU-1 and ICU-2. The 2 buildings were connected by a pedestrian bridge, and frequent movement of persons (medical workers, patients, and visitors) and portable medical devices occurred between the 2 buildings, providing many opportunities for the intrahospital transmission of bacterial pathogens between buildings and wards.
Average age of these patients was 72 years (range 39–94 years), and most (79%) were men. All but 1 KPC-3 isolates were detected >2 days after admission (range 4–328 days). KPC-3 E. coli isolate CG2126 (from patient 22) was detected from the blood sample of a patient on the same day of admission in April 2021; however, this patient had cholangiocarcinoma and had been hospitalized in the hepatobiliary and pancreatic surgery ward 2 weeks earlier. Most patients had serious underlying diseases, including type 2 diabetes mellitus (n = 8), hypertension (n = 4), hypoproteinemia (n = 4), and cerebral infarction (n = 2). Most patients had received β-lactam antimicrobial treatments, such as piperacillin/tazobactam (n = 17), meropenem (n = 12), tigecycline (n = 12), and cefoperazone/sulbactam (n = 9). Most patients (95.8%) underwent invasive procedures that involved medical devices, including attachment of a ventilator (23/24), deep vein intubation (9/24), and attachment of a urinary catheter (4/24) (Appendix 2 Table 1). In addition, most patients had prolonged hospital stays (range 8–462 days), including 6 patients who were hospitalized for >6 months (Figure 1, panel B). The prognosis of some patients was poor. Half of the patients (n = 12) had deteriorating health conditions during discharge, and 3 patients died during their hospital stay (Appendix 2 Table 1).
Starting in mid-2021 (and coinciding with the COVID-19 epidemic), the hospital enacted enhanced infection control measures, including chlorhexidine skin cleaning for ICU patients, improved hand hygiene compliance in healthcare workers, easy access to hand-hygiene supplies, restriction of hospital visitors, decontamination of the patients’ environment, and enhanced disinfection of medical equipment. Those measures also help to control the KPC-3 CRE outbreak, and only 1 carbapenem-resistant S. marcescens (attributable to NDM) was detected from August 2021 (data not shown).
Antimicrobial Susceptibility and Carbapenem Inactivation Assay
We then examined the susceptibility of the 25 isolates against 18 antibiotics (Appendix 2 Table 2). Our results indicated these isolates were all multidrug-resistant, exhibiting high-level resistance to all β-lactam antibiotics, including carbapenems, but remained susceptible to amikacin, gentamicin, and ceftazidime/avibactam (except CG2126). The E. hormaechei isolate CG2126 was resistant to ceftazidime/avibactam because of the co-existence of blaNDM-1 (Appendix 2 Table 1). Modified carbapenem inactivation method results confirmed that all 25 isolates were carbapenemase producers, consistent with the presence of blaKPC-3 (or blaNDM-1) genes among these isolates.
Genomic Phylogeny of KPC-3–producing Enterobacterales
We first conducted a core-genome phylogenetic analysis by using Parsnp (16) and compared our KPC-3–producing S. marcescens genomes with 748 S. marcescens genome assemblies from the National Center for Biotechnology Information RefSeq database (https://www.ncbi.nlm.nih.gov/refseq [accessed October 1, 2021]). A total of 73 strains were from China. The core-genome tree showed that the 18 KPC-3 S. marcescens strains formed a single cluster and were phylogenetically close to another cluster of 44 strains, which mostly harbored KPC-2 and were from China (named KPC-2 cluster) (Figure 2). Further core single-nucleotide polymorphism (SNP) distance analysis showed that the 18 outbreak strains differed by an average of 7 core SNPs (range 0–18), indicating clonal expansion. They differed from the China KPC-2 cluster strains by an average of 7,400 core SNPs (range 7,388–7,428) and differed from the remaining strains from China by an average of 45,082 core SNPs (range 11,173–58,247), suggesting that the KPC-3 strains belonged to a unique clone, which is consistent with the core-genome phylogeny (Figure 2).
Three K. pneumoniae strains carried blaKPC-3, and they belonged to 2 STs (ST65 and ST967), containing KL2 and KL18 type capsules. The 2 K2 ST65 strains were isolated from the same patient. ST65 K2 strains belonged to prototypical hypervirulent K. pneumoniae clone, harboring a battery of virulence genes, encoding yersiniabactin (ybt), colibactin (clb), aerobactin (iuc), and Salmochelin (iro). The 2 K2 strains also contained an IncHIB-FIB virulence plasmid, harboring the regulator of mucoid phenotype A genes rmpA and rmpA2. The 2 ST65 strains only differed by 2 core SNPs. The E. hormaechei strain CG2126 belonged ST127, and the E. coli strain was from a novel ST.
blaKPC-3–harboring IncX8 Plasmids
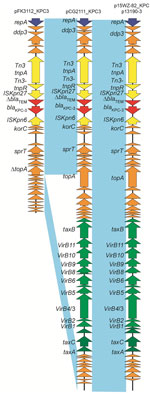
Two representative blaKPC-3-plasmids (pCG2111-KPC-3 and pFK3112-KPC-3) were completely sequenced. De novo assembly of the plasmid sequences generated a single head-to-tail contig for each plasmid. PlasmidFinder 2.1 assigned the 2 plasmids as the IncX5_2 (GenBank accession no. MF062700), whereas a recent study has reassigned IncX5_2 plasmids as a novel IncX8 group (17).
The pCG2111-KPC-3 was 41,852 bp in length, with an average G+C content of 46%, and harbored 57 predicted open reading frames, with blaKPC-3 the only intact antimicrobial resistance gene. A BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi) showed that pCG2111-KPC-3 was almost identical to plasmids p13190–3 (blaKPC-2–harboring; GenBank accession no. MF344555) (17), isolated from ST392 K. pneumoniae in 2013, and p15WZ-82_KPC (blaKPC-2–harboring; GenBank accession no. CP032355) (18), isolated from ST595 K. variicola in 2015 in China. In addition, both blaKPC-3 (in pCG2111-KPC-3) and blaKPC-2 (in p13190–3 and p15WZ-82_KPC) genes were carried by a conserved Tn3 transposon, with the structure of tnpA-npR-ISkpn27-ΔblaTEM-blaKPC-2/3-ISkpn6. We observed 2 major differences between pCG2111-KPC-3 and the other 2 Klebsiella IncX8 plasmids: first, pCG2111-KPC-3 harbors blaKPC-3, whereas the other 2 carry blaKPC-2; second, the 2 Klebsiella IncX8 plasmids have 8 22-bp iterons located upstream from the replication gene, whereas pCG2111-KPC-3 only has 7 copies of iteron, and 1 iteron (AAACATGATGATAAATGCGAAT) was deleted (Figure 3, 4).
The pFK3112-KPC-3 plasmid was smaller (21,888 bp in length) and had an average G+C content of 48%, carrying the same IncX8 replicon, and harbored 36 predicted open reading frames. In comparison to pCG2111-KPC-3, the only difference is that pFK3112-KPC-3 has a 19,964-bp deletion flanked by 5-bp repeat sequences of GCATC, encompassing the entire transfer operon from the type IA DNA topoisomerase gene top to the DNA distortion polypeptide gene taxA (Figure 3, 4).
We then used pCG2111-KPC-3 and pFK3112-KPC-3 as the reference sequences, and we used the reference mapping and mauve contig mover (19) functions in Geneious Prime 2020 (https://www.geneious.com) to reconstruct the IncX8 plasmids from the remaining 23 blaKPC-3–haroboring strains. The analysis showed that the S. marcescens (n = 18), E. coli (n = 1), E. hormaechei (n = 1) (CG2039), P. mirabilis (n = 1), and ST65 K2 K. pneumoniae strains (n = 2) carried pCG2111-KPC-3–like plasmids. One E. hormaechei isolate (CG2126) had the pFK3112-KPC-3–like plasmid, with a ≈20-kb deletion in comparison to pCG2111-KPC-3.
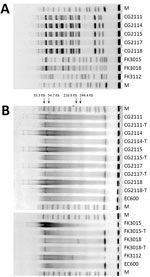
We then used conjugation assay to evaluate the transconjugation ability and frequency of blaKPC-3–harboring IncX8 plasmids. We selected 7 strains, 5 S. marcescens and 2 ST65 K2 K. pneumoniae, as the donors and the E. coli EC600 as the recipient strain. The blaKPC-3 plasmids from these isolates were all successfully transferred to the recipient strain. The S1-nuclease PFGE pattern showed that all the 7 transconjugants had only 1 plasmid, at a size of ≈42 kb (Figure 5). The transfer frequencies of the 7 strains ranged from 1.57 × 10−3 to 7.8 × 10−4. The antimicrobial susceptibility testing results further confirmed the carbapenem resistance had been transferred to recipient strains. In addition, we tested the transconjugation ability of pFK3112-KPC-3 in K. pneumoniae FK3112, and the result showed that pFK3112-KPC-3 failed to conjugate, which is consistent with the sequence analysis showing the lack of tra operon.
Ceftazidime/avibactam in vitro Selection Assays
Ceftazidime/avibactam has been increasingly used in China to treat CRE infections, especially those attributable to KPC producers (20,21). We conducted a subinhibitory concentration antimicrobial selection experiment in 24 KPC-3 strains (except CG2126) to examine their potential to develop ceftazidime/avibactam resistance. After induced selection by 1/2 MIC concentration of ceftazidime/avibactam, 22 isolates (all except 2 S. marcescens) developed resistance (MIC >16/4 μg/mL), and the resistant rate was as high as 91.7%. By contrast, we conducted the same in vitro selection in 24 blaKPC-2–harboirng K. pneumoniae strains. Only 1 strain developed ceftazidime/avibactam resistance (MIC >16/4 μg/mL) after 1/2 MIC induction, and the resistant rate was as low as 4.2%, which is significantly lower that of the induced resistance rate of KPC-3 strains (p<0.05). These results further suggested that these KPC-3-producing strains can be easily selected for resistance to ceftazidime/avibactam.
G. mellonella Infection Model of KPC-3–Producing Hypervirulent K2 K. pneumoniae Strains
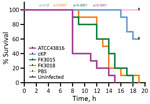
K. pneumoniae K2 ST65 strains belong to the prototypical hypervirulent clone. A string test of the ST65 strains FK3015 and FK3018 showed positive results, consistent with their genotypes. To assess the potential virulence of these 2 isolates, we conducted the G. mellonella larvae infection experiment (Figure 6). After 20 hours of infection, blaKPC-3–harboring ST65 strains and the hypervirulent reference K2 K. pneumoniae strain ATCC 43816 showed a 100% mortality rate, which was significantly higher than that observed in larvae infected with nontoxic reference classical K. pneumoniae strains (p<0.05). These results indicate that the KPC-3 K2 ST65 K. pneumoniae are highly virulent and the acquisition of blaKPC-3–harboring IncX8 plasmids does not comprise the virulence potential in these strains.
In this study, we report a KPC-3–producing Enterobacterales outbreak in China. In China, the spread of blaKPC-2 was primarily associated with IncFII(pHN7A8)-R plasmids and with epidemic K. pneumoniae ST11 strains (22–24), whereas this KPC-3 outbreak was primarily associated with the clonal expansion of S. marcescens and was mediated by an uncommon IncX8 plasmid. We also detected horizontal transmission of blaKPC-3–harboring IncX8 plasmids in different Enterobacterales species. The S. marcescens, E. coli, P. mirabilis, E. hormaechei (CG2126), and the K. pneumoniae ST65 strains harbored the same plasmid as pCG2111-KPC-3, indicating horizontal transmission of blaKPC-3 plasmids in these strains. However, the origin of pCG2111-KPC-3 remains unclear. Although KPC-3 was initially detected in an S. marcescens isolate (CG2008), the possibility that CG2008 acquired this plasmid from other strains cannot be ruled out. blaKPC-2 IncX8 plasmids have been reported in Klebsiella isolates in China, and in our study 2 KPC-3–producing K. pneumoniae strains were recovered nearly at the same time as CG2008 (Appendix 2 Table 1). In addition, S. marcescens strains might have obtained pCG2111-KPC-3–like plasmids from other strains (e.g., K. pneumoniae), which was then followed by clonal expansion.
The E. hormaechei strain (CG2039) in patient 24 and the K. pneumoniae ST967 strain (FK3112) from patient 20 harbored the same truncated plasmid (pFK3112-KPC-3), which may arise by recombination after the acquisition of intact plasmid. However, our conjugation experiment showed that pFK3112-KPC-3 cannot self-conjugate, and thus a possible explanation of the presence of pFK3112-KPC-3 in E. hormaechei and K. pneumoniae strain was that the same recombination happened independently in both species or pFK3112-KPC-3 was transferred with the assistance of helper plasmids. Our analysis demonstrated that plasmid-mediated horizontal and vertical transmission have played important roles in the KPC-3 Enterobacterales outbreak.
The blaKPC-3–harboring InX8 plasmid pCG2111-KPC-3 was almost identical to the blaKPC-2–harboring plasmids p13190–3 and p15WZ-82_KPC from K. pneumoniae and K. variicola strains in China, suggesting that KPC-3 probably originated through a single amino acid substitution on the same IncX8 plasmid. A similar KPC-2 to KPC-3 change has also been described in other blaKPC–harboring plasmids, including the epidemic pKpQIL-like plasmids (25). However, plasmids p13190–3 and p15WZ-82_KPC were identified in 7 (2013) and 5 (2015) years before the KPC-3 outbreak (2020), indicating that the blaKPC-2–harboring IncX8 plasmids have already existed and possibly circulated in China previously.
Compared with the predominant blaKPC-2–harboring IncFII (pHN7A8)-R plasmids (>100 kb) in China, the blaKPC-3–harboring IncX8 plasmid has a smaller genome size (≈42 kb). Our results showed the conjugation frequencies of blaKPC-3–harboring IncX8 ranged from 1.57 × 10−3 to 7.8 × 10−4 per donor cell, which is similar to that of the blaKPC-2–harboring plasmids (6.3 × 10−3 to 1 × 10−4) (26–28) and the epidemic blaNDM–harboring IncX3 plasmids (29,30), which have spread widely across different sectors of the human population, the animal population, and the environment (29–31). Compared with the previously reported blaKPC-2–harboring plasmids (p13190-3 and p15WZ-82_KPC), our KPC-3 IncX8 plasmids have 1 less copy of iterons in the replication origin. The iterons are essential for plasmid replication and inhibition of plasmid overreplication (32). Whether the deletion of an iteron copy could affect the plasmid replication or copy numbers, leading to increase plasmid transfer, is unclear, which warrants further studies. Nevertheless, our study clearly demonstrates that IncX8 plasmids can transfer across different clinical Enterobacterales species. Our G. mellonella infection model results also indicated that the acquisition of blaKPC-3–harboring IncX8 in clinical hypervirulent ST65 K2 K. pneumoniae strains does not lead to reduced virulence. This observation could be another example of the emergence of carbapenem-resistant and hypervirulent K. pneumoniae strains attributable to the horizontal transfer of blaKPC-3–harboring IncX8 plasmids into prototypically hypervirulent K2 strains.
Although most KPC-3 producing strains were multidrug-resistant, most of them remained sensitive to ceftazidime/avibactam. In China, ceftazidime/avibactam has been approved for clinical treatment since 2019. However, resistance emerged soon after the clinical use of ceftazidime/avibactam in different regions, including China; this resistance usually was associated with mutations in the omega loop of KPC enzymes (33–35). Ceftazidime/avibactam resistance appeared to occur more frequently in a KPC-3 than a KPC-2 background, presumably because of the higher hydrolysis activity of KPC-3 against ceftazidime (8,36). Our subinhibitory concentrations ceftazidime/avibactam selection experiment results showed that most of our KPC-3–producing strains developed resistance, and the resistance rate was as high as 91.7%. By contrast, the KPC-2-producing strains showed a <4.2% rate for developing ceftazidime/avibactam resistance. However, PCR and Sanger sequencing of subinhibitory concentrations ceftazidime/avibactam–selected KPC-3 strains (3 colonies of each stain) failed to identify amino acid mutation in KPC-3 (data not shown). We suspected alternative mechanism, such as the increased gene copy numbers, expressions, or both (37,38), might contribute to ceftazidime/avibactam resistance. Nevertheless, our results suggested that the IncX8 blaKPC-3 strains may readily develop ceftazidime/avibactam resistance during treatment, despite being susceptible in vitro, which poses a major challenge for the clinical application of ceftazidime/avibactam as a last resort for treating CRE infections.
In this study, most patients had underlying diseases, had lengthy hospital stays, and underwent invasive medical device treatments (e.g., treatments involving a ventilator). Mechanical ventilation is a known risk factor of nosocomial infections, including CRE-attributable infections. The close proximity of these medical wards and movement of persons between the 2 buildings probably promoted the spread of KPC-3 strains between different wards. However, this outbreak went unrecognized and unconfirmed during routine surveillance, until our genomic study commenced in later 2021. Nevertheless, the COVID-19–related enhanced infection control measures already in place effectively controlled the KPC-3 outbreak. Unfortunately, this outbreak was not recognized and confirmed during routine surveillance, which should have detected the high numbers of multidrug-resistant Serratia infections as an unusual, and possibly epidemic, occurrence. Our results further emphasize that genomic surveillance and improved infection control practice are essential to tackle hospital outbreaks.
In summary, we report a KPC-3 Enterobacterales outbreak in China, which involved both clonal and horizontal transmissions of carbapenem resistance. The further spread of the blaKPC-3–harboring IncX8 plasmids and these KPC-3 strains in China and other global regions should be closely monitored.
Ms. Chen is a researcher at Ningbo First Hospital Department of Clinical Laboratory Medicine whose research interests include antibiotic-resistance mechanisms. Ms. Ai is a researcher at First Affiliated Hospital of Wenzhou Medical University Department of Respiratory Medicine whose research interests include antibiotic-resistance mechanisms. Dr. Zhou is a researcher at Shanghai Pulmonary Hospital Department of Clinical Laboratory Medicine, Tongji University School of Medicine whose research interests include antibiotic-resistance mechanisms.
Acknowledgment
This study was supported by grants from Shanghai Pulmonary Hospital’s Department of Clinical Laboratory Medicine and the Shanghai Sailing Program (grant no. 22YF1437500).
References
- Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother. 2009;53:3365–70. DOIPubMedGoogle Scholar
- Piazza A, Principe L, Comandatore F, Perini M, Meroni E, Mattioni Marchetti V, et al. Whole-genome sequencing investigation of a large nosocomial outbreak caused by ST131 H30Rx KPC-producing Escherichia coli in Italy. Antibiotics (Basel). 2021;10:718. DOIPubMedGoogle Scholar
- Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. DOIPubMedGoogle Scholar
- Hirsch EB, Tam VH. Detection and treatment options for Klebsiella pneumoniae carbapenemases (KPCs): an emerging cause of multidrug-resistant infection. J Antimicrob Chemother. 2010;65:1119–25. DOIPubMedGoogle Scholar
- Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22:686–96. DOIPubMedGoogle Scholar
- Zhang Y, Wang Q, Yin Y, Chen H, Jin L, Gu B, et al. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE Network. Antimicrob Agents Chemother. 2018;62:e01882–17. DOIPubMedGoogle Scholar
- Zhang R, Liu L, Zhou H, Chan EW, Li J, Fang Y, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. DOIPubMedGoogle Scholar
- Alba J, Ishii Y, Thomson K, Moland ES, Yamaguchi K. Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing beta-lactamase. Antimicrob Agents Chemother. 2005;49:4760–2. DOIPubMedGoogle Scholar
- Li G, Wei Q, Wang Y, Du X, Zhao Y, Jiang X. Novel genetic environment of the plasmid-mediated KPC-3 gene detected in Escherichia coli and Citrobacter freundii isolates from China. Eur J Clin Microbiol Infect Dis. 2011;30:575–80. DOIPubMedGoogle Scholar
- Du H, Chen L, Chavda KD, Pandey R, Zhang H, Xie X, et al. Genomic characterization of Enterobacter cloacae isolates from China that coproduce KPC-3 and NDM-1 carbapenemases. Antimicrob Agents Chemother. 2016;60:2519–23. DOIPubMedGoogle Scholar
- Miao M, Wen H, Xu P, Niu S, Lv J, Xie X, et al. Genetic diversity of carbapenem-resistant (CRE) clinical isolates from a tertiary hospital in eastern China. Front Microbiol. 2018;9:3341. DOIPubMedGoogle Scholar
- Wei J, Zou C, Wang D, Huang A, Niu S. Genetic diversity and in vitro activity of ceftazidime/avibactam and aztreonam/avibactam against imipenem-resistant Enterobacteriaceae isolates in Southwest China: A single-centre study. J Glob Antimicrob Resist. 2020;22:448–51. DOIPubMedGoogle Scholar
- Tang H-J, Chen Y-T, Chiang T, Fung C-P, Chuang Y-C, Kristopher Siu L. Identification of the first imported KPC-3 Klebsiella pneumoniae from the USA to Taiwan. Int J Antimicrob Agents. 2014;44:431–5. DOIPubMedGoogle Scholar
- Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–58. DOIPubMedGoogle Scholar
- Nordmann P, Poirel L, Carrër A, Toleman MA, Walsh TR. How to detect NDM-1 producers. J Clin Microbiol. 2011;49:718–21. DOIPubMedGoogle Scholar
- Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. DOIPubMedGoogle Scholar
- Fang H, Feng J, Xu Y, Zhang Y, Zhan Z, Yin Z, et al. Sequencing of pT5282-CTXM, p13190-KPC and p30860-NR, and comparative genomics analysis of IncX8 plasmids. Int J Antimicrob Agents. 2018;52:210–7. DOIPubMedGoogle Scholar
- Yang X, Wai-Chi Chan E, Zhang R, Chen S. A conjugative plasmid that augments virulence in Klebsiella pneumoniae. Nat Microbiol. 2019;4:2039–43. DOIPubMedGoogle Scholar
- Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics. 2009;25:2071–3. DOIPubMedGoogle Scholar
- Tumbarello M, Trecarichi EM, Corona A, De Rosa FG, Bassetti M, Mussini C, et al. Efficacy of ceftazidime-avibactam salvage therapy in patients with infections caused by Klebsiella pneumoniae carbapenemase–producing K. pneumoniae. Clin Infect Dis. 2019;68:355–64. DOIPubMedGoogle Scholar
- van Duin D, Bonomo RA. Ceftazidime/avibactam and ceftolozane/tazobactam: second-generation β-lactam/β-lactamase inhibitor combinations. Clin Infect Dis. 2016;63:234–41. DOIPubMedGoogle Scholar
- Shen P, Zhang Y, Li G, Jiang X. Characterization of the genetic environment of the blaKPC-2 gene among Klebsiella pneumoniae isolates from a Chinese Hospital. Braz J Infect Dis. 2016;20:384–8. DOIPubMedGoogle Scholar
- Xu J, Zhao Z, Ge Y, He F. Rapid emergence of a pandrug-resistant ST11 isolate in an inpatient in a teaching hospital in China after treatment with multiple broad-spectrum antibiotics. Infect Drug Resist. 2020;13:799–804. DOIPubMedGoogle Scholar
- Zhou H, Zhang K, Chen W, Chen J, Zheng J, Liu C, et al. Epidemiological characteristics of carbapenem-resistant collected from 17 hospitals in Nanjing district of China. Antimicrob Resist Infect Control. 2020;9:15. DOIPubMedGoogle Scholar
- Chen L, Chavda KD, Melano RG, Jacobs MR, Koll B, Hong T, et al. Comparative genomic analysis of KPC-encoding pKpQIL-like plasmids and their distribution in New Jersey and New York Hospitals. Antimicrob Agents Chemother. 2014;58:2871–7. DOIPubMedGoogle Scholar
- Li JJ, Lee CS, Sheng JF, Doi Y. Complete sequence of a conjugative incn plasmid harboring blaKPC-2, blaSHV-12, and qnrS1 from an Escherichia coli sequence type 648 strain. Antimicrob Agents Chemother. 2014;58:6974–7. DOIPubMedGoogle Scholar
- Liu X, Chan EW-C, Chen S. Transmission and stable inheritance of carbapenemase gene (blaKPC-2 or blaNDM-1)-encoding and mcr-1-encoding plasmids in clinical Enterobacteriaceae strains. J Glob Antimicrob Resist. 2021;26:255–61. DOIPubMedGoogle Scholar
- Papagiannitsis CC, Bitar I, Malli E, Tsilipounidaki K, Hrabak J, Petinaki E. IncC blaKPC-2-positive plasmid characterised from ST648 Escherichia coli. J Glob Antimicrob Resist. 2019;19:73–7. DOIPubMedGoogle Scholar
- Zhao J, Zhang Y, Fan Y, Han J, Xiong Z, Liu X, et al. Characterization of an NDM-5-producing hypervirulent sequence type 65 clone from a lung transplant recipient. Emerg Microbes Infect. 2021;10:396–9. DOIPubMedGoogle Scholar
- Kong LH, Lei CW, Ma SZ, Jiang W, Liu BH, Wang YX, et al. Various sequence types of Escherichia coli isolates coharboring blaNDM-5 and mcr-1 genes from a commercial swine farm in China. Antimicrob Agents Chemother. 2017;61:e02167–16. DOIPubMedGoogle Scholar
- Sugawara Y, Akeda Y, Hagiya H, Sakamoto N, Takeuchi D, Shanmugakani RK, et al. Spreading patterns of NDM-producing in clinical and environmental settings in Yangon, Myanmar. Antimicrob Agents Chemother. 2019;3:63. DOIPubMedGoogle Scholar
- Konieczny I, Bury K, Wawrzycka A, Wegrzyn K. Iteron Plasmids. Microbiol Spectr. 2014;2:
2.6.14 . DOIPubMedGoogle Scholar - Winkler ML, Papp-Wallace KM, Bonomo RA. Activity of ceftazidime/avibactam against isogenic strains of Escherichia coli containing KPC and SHV β-lactamases with single amino acid substitutions in the Ω-loop. J Antimicrob Chemother. 2015;70:2279–86. DOIPubMedGoogle Scholar
- Hemarajata P, Humphries RM. Ceftazidime/avibactam resistance associated with L169P mutation in the omega loop of KPC-2. J Antimicrob Chemother. 2019;74:1241–3. DOIPubMedGoogle Scholar
- Haidar G, Clancy CJ, Shields RK, Hao B, Cheng S, Nguyen MH. Mutations in blaKPC-3 that confer ceftazidime-avibactam resistance encode novel KPC-3 variants that function as extended-spectrum β-lactamases. Antimicrob Agents Chemother. 2017;61:e02534–16. DOIPubMedGoogle Scholar
- Mehta SC, Rice K, Palzkill T. Natural variants of the KPC-2 carbapenemase have evolved increased catalytic efficiency for ceftazidime hydrolysis at the cost of enzyme stability. PLoS Pathog. 2015;11:
e1004949 . DOIPubMedGoogle Scholar - Coppi M, Di Pilato V, Monaco F, Giani T, Conaldi PG, Rossolini GM. Ceftazidime-avibactam resistance associated with increased blaKPC-3 gene copy number mediated by pKpQIL plasmid derivatives in sequence type 258 Klebsiella pneumoniae. Antimicrob Agents Chemother. 2020;64:e01816–9. DOIPubMedGoogle Scholar
- Nelson K, Hemarajata P, Sun D, Rubio-Aparicio D, Tsivkovski R, Yang S, et al. Resistance to ceftazidime-avibactam is due to transposition of KPC in a porin-deficient strain of Klebsiella pneumoniae with increased efflux activity. Antimicrob Agents Chemother. 2017;61:e00989–17. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: June 08, 2022
1These first authors contributed equally to this article.
Table of Contents – Volume 28, Number 7—July 2022
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Fangyou Yu, Shanghai Pulmonary Hospital Department of Clinical Laboratory Medicine, Tongji University School of Medicine, No. 507, Zhengmin Rd, Yangpu District, Shanghai, China; e-mail:wzjxyfy@163.com ; Liang Chen, Hackensack Meridian Health Center for Discovery and Innovation, Center for Discovery and Innovation, 111 Ideation Way, Nutley, NJ 07110, USA
Top