Volume 28, Number 7—July 2022
Research
Analyzing and Modeling the Spread of SARS-CoV-2 Omicron Lineages BA.1 and BA.2, France, September 2021–February 2022
Figure 5
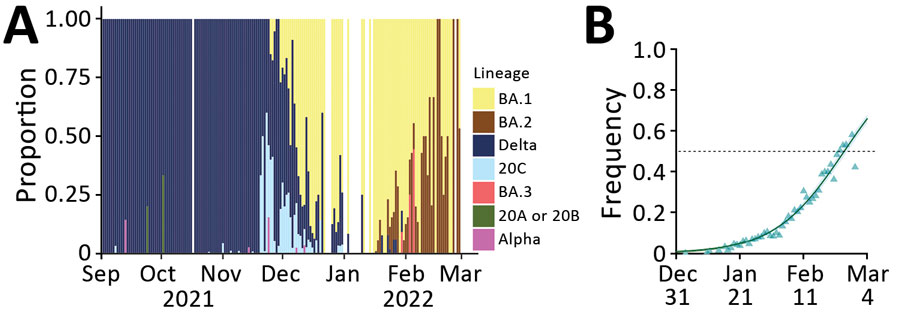
Figure 5. Monitoring and quantifying SARS-CoV-2 variant spread using whole-genome sequencing, France. A) Raw proportion of SARS-CoV-2 lineages inferred from whole-genome sequences of 16,973 samples. B) Estimated proportion and growth advantage of the BA.2 variant with respect to the BA.1 variant. Raw occurrence data from panel A is stratified by region in Appendix 1 Figure).
Page created: May 03, 2022
Page updated: June 18, 2022
Page reviewed: June 18, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.