Volume 28, Number 9—September 2022
Dispatch
International Spread of Multidrug-Resistant Rhodococcus equi
Cite This Article
Citation for Media
Abstract
A multidrug-resistant clone of the animal and human pathogen Rhodococcus equi, MDR-RE 2287, has been circulating among equine farms in the United States since the 2000s. We report the detection of MDR-RE 2287 outside the United States. Our finding highlights the risk for MDR-RE spreading internationally with horse movements.
Rhodococcus equi is a soilborne aerobic actinomycete bacterium that infects animals and humans. Human infections are opportunistic, zoonotic in origin, and considered to be linked to exposure to farm environments (1–3). Although clinical R. equi infections are relatively rare in most animal species, foals are commonly affected, and incidence is often high in horse farms in equine breeding countries (4). Long courses of rifampin and a macrolide is the mainstay therapy for foal rhodococcosis. This treatment has been systematically used since the 1980s, and no significant resistance was detected until the early 2000s, after mass prophylactic application at R. equi–endemic farms in the United States (5,6).
The emerging dual macrolide–rifampin resistance is attributable to a multidrug-resistant R. equi (MDR-RE) clone, named 2287, which has spread among horse farms across the United States. MDR-RE 2287 arose by co-acquisition of the conjugative plasmid pRErm46 and a specific rpoBS531F (TCG to TTC) mutation conferring high-level rifampin resistance (7,8). pRErm46 specifies resistance to macrolides, lincosamides, and streptogramins via the erm(46) gene carried on TnRErm46, a highly mobile transposon, and to sulfonamides, streptomycin, spectinomycin, tetracycline, and doxycycline via a class 1 integron (C1I) and associated tetRA(33) determinant (9). Although it has so far been found only in the United States, we predicted MDR-RE could spread to other countries with the movement of equids (10).
After MDR-RE was characterized in 2019 (8), we established an informal surveillance network with colleagues in North and South America, Europe, the United Kingdom, Africa, Asia, and Australia. We asked collaborating laboratories to review their retrospective R. equi collections and prospectively identify isolates with erythromycin MIC >4 μg/mL, potentially denoting erm(46)-mediated macrolide resistance. Two equine clinical strains from necropsied foals in Ireland met the criterion: PAM2528, recovered in 2016, and PAM2578, recovered in 2021 (henceforth designated as 2528 and 2578). Both strains originated from the same farm and had MICs >32 μg/mL for erythromycin and >256 μg/mL for rifampin, consistent with MDR-RE’s resistance phenotype (7–10).
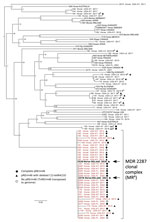
We confirmed that both isolates were erm(46) positive by PCR and carried the rpoBS531F mutation unique to the MDR-RE 2287 clone using previously described methods (9). A PCR using primers 5′-CCGAGATGTGTCGGACTTC-3′ (forward) and 5′-CGCCGAAGAACAACCCGAGGATG-3′ (reverse) showed the pRErm46 resistance plasmid carried the ΔC1I-tetRA deletion, observed in some recent MDR-RE isolates (9,10). Accordingly, 2528 and 2578 were susceptible to trimethoprim/sulfamethoxazole, streptomycin, spectinomycin, and tetracycline, to which the C1I-tetRA(33) determinant confers resistance (9). We used paired-end Illumina sequencing (Illumina, https://www.illumina.com) to obtain genomic libraries of the isolates. Reads were quality-checked using FastQC version 0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc), trimmed with TrimmomaticPE version 0.39 (11), and assembled using SPAdes v3.15.2 (12). For 2528 sequences, average forward/reverse Phred score was 38/38, coverage depth 79×, and number of contigs (>1 kb) 117. For 2578, average Phred score was 36/36; coverage depth, 243×; and number of contigs, 26. BlastN searches (https://blast.ncbi.nlm.nih.gov/Blast.cgi) confirmed presence of pRErm46 sequences in the draft genomes. We used ParSNP version 1.5.6 (13) and FastTree version 2.1.11 (http://www.microbesonline.org/fasttree) to build approximate maximum-likelihood trees based on core single-nucleotide polymorphisms (SNPs) to determine the position of the isolates in the R. equi population structure. The output R. equi tree showed that the 2 resistant isolates from Ireland belonged to the MDR-RE 2287 clone (Figure 1).
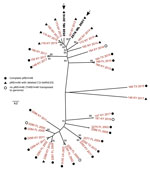
Because the short genetic distances compressed the branching of the MDR-RE 2287 isolates, we repeated the phylogenetic analysis with only the clonal genomes to explore their relationships in more detail (Figure 2). For this analysis, we detected core SNPs using SNIPPY version 4.6.0 (https://github.com/tseemann/snippy), which avoided genome alignment errors observed with ParSNP that distort the phylogenetic reconstruction of virtually identical isolates.
The consensus maximum-likelihood tree subdivided the MDR-RE 2287 clonal complex in 2 main sublineages, one comprising older isolates from 2002–2011 and the other isolates from 2015 and later. R. equi 2528 and 2578 were located in 2 adjacent top branches within the younger sublineage, together with all (n = 7) isolates from New York. Because the analyzed MDR-RE 2287 collection comprised isolates from different US locations, the clustering with the New York isolates suggested a common origin. The New York isolates were recovered over a period of several years since 2012, pointing to a locally circulating MDR-RE 2287 subpopulation as the likely source of the isolates later found in Ireland. This possibility was further supported by the finding that both the 2528 and 2578 genomes possessed ΔC1I-tetRA pRErm46 variants, also carried by the New York subpopulation but only exceptionally by other MDR-RE 2287 complex members (Figures 1, 2).
We document the international spread of the MDR-RE 2287 clone that has been circulating in the United States since the 2000s (8,10). MDR-RE 2287 appears to be following the same pattern of the pandemic multidrug-resistant clones of human bacterial pathogens, which within a few years after emergence and initial local expansion become globally disseminated (15). For MDR-RE, the process is slower, likely because of fewer opportunities for transmission afforded by horse trade and contacts compared with the scale of human interactions and travel.
The positioning of the Ireland isolates in 2 separate subbranches of the New York radiation (Figure 2) may indicate they represent independent, temporally distinct import events that took place around 2016 and 2021, involving different MDR-RE 2287 subclones. That MDR-RE 2287 was not detected again in Ireland until 5 years later indicates that it might not have persisted after its first appearance in the country in 2016. This scenario may be explained by the different R. equi–targeted equine farm management in Ireland compared with that in the United States, where the emergence, maintenance, and spread of MDR-RE was favored by the application of mass antibioprophylaxis at R. equi–endemic farms (6,8). This practice was not implemented on the affected farm, nor is it applied in Ireland in general. However, the genetic distance between the 2528 and 2578 strains is comparable to that for the 7 New York isolates, recovered during 2012–2015. It cannot therefore be excluded that the 2 Ireland strains represent successive isolations of a locally evolving single imported MDR-RE 2287 subclone (21 SNPs difference over 5M bp, ≈1 × 10−6 substitutions/site/year, consistent with normal genetic drift values).
A Kentucky isolate of the ΔC1I-tetRA type was also located in a terminal branch of the New York cluster, whereas Kentucky isolates with complete pRErm46 plasmids were positioned at basal bifurcations of the radiation (e.g., the 148, 152, and 153 clusters) (Figure 2). These observations suggest a transmission history in which a relatively recent MDR-RE 2287 subclone that acquired a pRErm46 ΔC1I-tetRA deletion, possibly originating from Kentucky where MDR-RE emerged and is prevalent (8,9), became endemic in a New York farm and was transferred, directly or indirectly, to Ireland. International trade in thoroughbred horses is frequent, and the affected farm in Ireland received horses from the United States, Europe, and the United Kingdom on a regular basis. Previous phylogenomic studies provided evidence of global circulation of R. equi genomotypes, probably linked to livestock trade (14). Our findings reinforce this notion and warn about the risk for MDR-RE becoming globally disseminated over time with horse movements.
Our study is not comprehensive but based on the voluntary collaboration of a small number of international colleagues, and thus MDR-RE may have also spread to other countries. We recommend actively monitoring the occurrence of the emerging MDR-RE 2287 clone for which, as our data highlight, erm(46) and the rpoBS531F (TCG→TTC) mutation can be used as molecular markers, eventually complemented with pRErm46/ΔC1I-tetRA(33) variant detection.
Dr. Val-Calvo is a postdoctoral fellow at the Laboratory of Microbial Pathogenesis, Medical School of the University of Edinburgh. His primary research interests include molecular microbiology, plasmid biology, bacterial genomics and evolution, and antimicrobial resistance.
Acknowledgments
We thank the network of international collaborators for their participation in macrolide- and rifampin-resistant R. equi monitoring.
New R. equi genome assemblies were deposited in GenBank under accession nos. JAJNNF000000000 (PAM2528) and JAJNNG000000000 (PAM2578).
Work on multidrug-resistant R. equi at J.V.-B.’s laboratory is supported by HBLB (grant no. prj-796).
References
- Prescott JF. Rhodococcus equi: an animal and human pathogen. Clin Microbiol Rev. 1991;4:20–34. DOIPubMedGoogle Scholar
- Yamshchikov AV, Schuetz A, Lyon GM. Rhodococcus equi infection. Lancet Infect Dis. 2010;10:350–9. DOIPubMedGoogle Scholar
- Vázquez-Boland JA, Meijer WG. The pathogenic actinobacterium Rhodococcus equi: what’s in a name? Mol Microbiol. 2019;112:1–15. DOIPubMedGoogle Scholar
- Muscatello G, Leadon DP, Klayt M, Ocampo-Sosa A, Lewis DA, Fogarty U, et al. Rhodococcus equi infection in foals: the science of ‘rattles’. Equine Vet J. 2007;39:470–8. DOIPubMedGoogle Scholar
- Giguère S. Treatment of infections caused by Rhodococcus equi. Vet Clin North Am Equine Pract. 2017;33:67–85. DOIPubMedGoogle Scholar
- Burton AJ, Giguère S, Sturgill TL, Berghaus LJ, Slovis NM, Whitman JL, et al. Macrolide- and rifampin-resistant Rhodococcus equi on a horse breeding farm, Kentucky, USA. Emerg Infect Dis. 2013;19:282–5. DOIPubMedGoogle Scholar
- Anastasi E, Giguère S, Berghaus LJ, Hondalus MK, Willingham-Lane JM, MacArthur I, et al. Novel transferable erm(46) determinant responsible for emerging macrolide resistance in Rhodococcus equi. J Antimicrob Chemother. 2015;70:3184–90.PubMedGoogle Scholar
- Álvarez-Narváez S, Giguère S, Anastasi E, Hearn J, Scortti M, Vázquez-Boland JA. Clonal confinement of a highly mobile resistance element driven by combination therapy in Rhodococcus equi. MBio. 2019;10:e02260–19. DOIPubMedGoogle Scholar
- Erol E, Scortti M, Fortner J, Patel M, Vázquez-Boland JA. Antimicrobial resistance spectrum conferred by pRErm46 of emerging macrolide (multidrug)-resistant Rhodococcus equi. J Clin Microbiol. 2021;59:
e0114921 . DOIPubMedGoogle Scholar - Álvarez-Narváez S, Giguère S, Cohen N, Slovis N, Vázquez-Boland JA. Spread of multidrug-resistant Rhodococcus equi, United States. Emerg Infect Dis. 2021;27:529–37. DOIPubMedGoogle Scholar
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. DOIPubMedGoogle Scholar
- Anastasi E, MacArthur I, Scortti M, Alvarez S, Giguère S, Vázquez-Boland JA. Pangenome and phylogenomic analysis of the pathogenic actinobacterium Rhodococcus equi. Genome Biol Evol. 2016;8:3140–8. DOIPubMedGoogle Scholar
- Baker S, Thomson N, Weill FX, Holt KE. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science. 2018;360:733–8. DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: August 11, 2022
Table of Contents – Volume 28, Number 9—September 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
José A. Vázquez-Boland, Microbial Pathogenesis Laboratory, University of Edinburgh Medical School (Biomedical Sciences—Infection Medicine), Edinburgh BioQuarter, Chancellor's Building, 49 Little France Crescent, Edinburgh, UK EH16 4SB, UK
Top