Volume 29, Number 11—November 2023
Research Letter
Genomic Sequencing Surveillance to Identify Respiratory Syncytial Virus Mutations, Arizona, USA
Cite This Article
Citation for Media
Abstract
We conducted surveillance of respiratory syncytial virus (RSV) genomic sequences for 100 RSV-A and 27 RSV-B specimens collected during November 2022–April 2023 in Arizona, USA. We identified mutations within prefusion F-protein antigenic sites in both subtypes. Continued genomic surveillance will be critical to ensure RSV vaccine effectiveness.
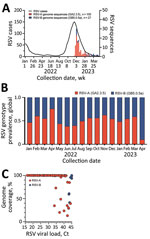
Respiratory syncytial virus (RSV), an RNA virus of the family Pneumoviridae, causes acute respiratory infections, primarily in children, adults with severe lung disease, and elderly persons (1). The United States experienced an early surge in RSV cases during the 2022–2023 respiratory pathogen season, coinciding with high levels of influenza and SARS-CoV-2 infections (2). In Arizona, USA, numbers of laboratory-confirmed RSV cases increased beginning in September 2022, peaked in mid-November 2022, and then declined to average levels by March 2023 (Figure, panel A).
The 2 major RSV subtypes, RSV-A and RSV-B, have distinct antigenic characteristics in the P, N, F, and G proteins (1). Each subtype is classified into genotypes based on sequence variability in the G protein (3). According to sequencing data from the GISAID database (4), global distribution of RSV genotypes during 2022–2023 was split between the GA2.3.5 genotype of RSV-A and the GB5.0.5a genotype of RSV-B (Figure, panel B). However, RSV vaccines are based on the prefusion conformation of the F protein. Because surveillance efforts should focus on whole-genome sequencing to better understand the evolution of the virus and its potential effect on vaccine efficacy, we conducted surveillance of genomic sequences from RSV-A and -B subtypes to identify genetic mutations. This study was approved by the Arizona State University Institutional Review Board (STUDY00011967).
We tested 127 RSV-positive nasopharyngeal swabs from previous standard-of-care respiratory pathogen testing at Valleywise Health Medical Center, which serves Maricopa County, Arizona, USA. Patients were a median of 22 years of age (interquartile range 2–44 years). We performed genomic surveillance to determine RSV strains circulating in Arizona during the 2022–2023 season. We performed 2 × 150-bp paired-end next-generation sequencing using a hybrid capture enrichment panel (Illumina Respiratory Virus Oligo Panel version 2, https://www.illumina.com). We used Trim Galore version 0.6.10 (https://github.com/FelixKrueger/TrimGalore) to quality filter and adaptor trim sequencing reads, mapped the reads to RSV-A and RSV-B reference sequences (GISAID accession nos. EPI_ISL_412866 and EPI_ISL_165399) using Burrows-Wheeler Aligner version 0.7.17-r1188 (https://bio-bwa.sourceforge.net), and generated consensus sequences using SAMtools version 1.17 (https://github.com/samtools/samtools/releases). We assembled 92 RSV-A (GA2.3.5 genotype) and 24 RSV-B (GB5.0.5a genotype) complete genome sequences and 8 RSV-A (GA2.3.5) and 3 RSV-B (GB5.0.5a) partial genome sequences (GenBank accession nos. OR143134–250; GISAID accession nos. EPI_ISL_17808760–814) (Figure, panel A).
To determine RSV viral load, we performed quantitative reverse transcription PCR using pan-HRSV assays that recognize both subtypes RSV-A and RSV-B (5). The mean RSV cycle threshold (Ct) value was 29.83 (SD 7.44). We found that specimens with viral load Ct ≤33 yielded 99%–100% genome coverage (Figure, panel C). Whole-genome phylogenetic analysis (Nextclade version 2.14.1, https://clades.nextstrain.org) showed that RSV-A sequences from Arizona clustered in clade GA2.3.5, indicating >3 independent introductions of RSV-A into Arizona (Appendix Figure 1). RSV-B sequences from Arizona formed a monophyletic group in clade GB5.0.5a, indicating a single introduction of strains locally transmitted within the state (Appendix Figure 1). Our findings were consistent with RSV investigations in Massachusetts (6) and Washington (7), USA, both of which suggested the atypical increase in cases during the 2022–2023 season resulted from multiple introductions of extant lineages, not a divergent RSV lineage with increased virulence or transmissibility.
The US Food and Drug Administration has approved 2 RSV vaccines for persons >60 years of age, both based on the RSV prefusion F protein. Arexvy (GlaxoSmithKline, https://www.gsk.com) is monovalent and Abrysvo (Pfizer, https://www.pfizer.com), bivalent (8,9). Most host antibodies target 6 antigenic sites (Ø–V) of the F protein (10). Within the F-gene sequences from RSV genomes from Arizona, we identified 7 nonsynonymous substitutions in antigenic sites I, II, IV, and V of RSV-A and 5 nonsynonymous substitutions in antigenic sites Ø, I, II, and V of RSV-B. We found each RSV-A genome mutation in <10% of samples. Most RSV-B genome mutations were more frequent; we found only 1 rare single-nucleotide polymorphism. Mutation frequencies were comparable with trends in recent global RSV genome sequences (Table). RSV-B S190N, S211N, and S389P mutations specifically have become increasingly dominant since 2020. Mutability at residue 389 was shared between subtypes, and each subtype had a preferential amino acid. Finally, we located the mutated residues on the prefusion F-protein crystal structure (protein data bank 7KQD). Many mutated residues were exposed on the F protein surface, suggesting they might interfere with antibody recognition (Appendix Figure 2).
Although RSV remains a substantial clinical burden, approved RSV vaccines reduce the risk of lower respiratory tract illness. By tracking RSV evolution, we can improve design of vaccine formulations to improve effectiveness. Our study was limited because we lacked understanding of potential functional consequences from mutations in F-protein antigenic sites. Although the G protein is under greater selective pressure and has higher mutation rates (3), observing its evolutionary trajectory in context with the F protein will be critical. Our study demonstrates the value of using whole-genome sequencing to identify genetic mutations in respiratory pathogens, including RSV, to ensure ongoing effectiveness of RSV vaccines.
Mrs. Holland is a research specialist at Arizona State University under the supervision of Dr. Efrem Lim. Her primary research interests are viral diseases and infections and how they influence viromes and microbiomes in human health.
Acknowledgments
We gratefully acknowledge Sarah Namdarian for assistance with collecting clinical specimens; Alexis Thomas, Gabrielle Hernandez Barrera, Michelle Tan for assistance with specimen processing; and Regan Sullins for assistance with constructing the resource library. We thank the authors from originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared through GISAID (https://www.gisaid.org) and GenBank.
This study was supported in part by Arizona State University and the Centers for Disease Control and Prevention (CDC BAA 75D30121C11084).
Contributions: conceptualization: E.S.L.; formal analysis: L.A.H., S.C.H., M.F.S., V.R.L.; investigation: L.A.H., S.C.H., M.F.S., V.R.L., E.S.L.; resources: V.M., L.N., M.M., R.S., M.W.; data curation: L.A.H., S.C.H., M.F.S., V.R.L.; writing, original draft: L.A.H., S.C.H., M.F.S., E.S.L.; writing, review and editing: L.A.H., S.C.H., M.F.S., L.N., E.S.L.; supervision: E.S.L.; funding acquisition: E.S.L. All authors reviewed and approved the final manuscript.
References
- Tin Tin Htar M, Yerramalla MS, Moïsi JC, Swerdlow DL. The burden of respiratory syncytial virus in adults: a systematic review and meta-analysis. Epidemiol Infect. 2020;148:
e48 . DOIPubMedGoogle Scholar - Centers for Disease Control and Prevention. Increased respiratory virus activity, especially among children, early in the 2022–2023 fall and winter [cited 2023 Jun 16]. https://emergency.cdc.gov/han/2022/han00479.asp#print
- Muñoz-Escalante JC, Comas-García A, Bernal-Silva S, Robles-Espinoza CD, Gómez-Leal G, Noyola DE. Respiratory syncytial virus A genotype classification based on systematic intergenotypic and intragenotypic sequence analysis. Sci Rep. 2019;9:20097. DOIPubMedGoogle Scholar
- Khare S, Gurry C, Freitas L, Schultz MB, Bach G, Diallo A, et al. GISAID’s role in pandemic response. China CDC Wkly. 2021;3:1049–51. DOIPubMedGoogle Scholar
- Wang L, Piedra PA, Avadhanula V, Durigon EL, Machablishvili A, López MR, et al. Duplex real-time RT-PCR assay for detection and subgroup-specific identification of human respiratory syncytial virus. J Virol Methods. 2019;271:
113676 . DOIPubMedGoogle Scholar - Adams G, Moreno GK, Petros BA, Uddin R, Levine Z, Kotzen B, et al. Viral lineages in the 2022 RSV surge in the United States. N Engl J Med. 2023;388:1335–7. DOIPubMedGoogle Scholar
- Goya S, Sereewit J, Pfalmer D, Nguyen TV, Bakhash SAKM, Sobolik EB, et al. Genomic characterization of respiratory syncytial virus during 2022–23 outbreak, Washington, USA. Emerg Infect Dis. 2023;29:865–8. DOIPubMedGoogle Scholar
- Papi A, Ison MG, Langley JM, Lee D-G, Leroux-Roels I, Martinon-Torres F, et al.; AReSVi-006 Study Group. AReSVi-006 Study Group. Respiratory syncytial virus prefusion F protein vaccine in older adults. N Engl J Med. 2023;388:595–608. DOIPubMedGoogle Scholar
- Kampmann B, Madhi SA, Munjal I, Simões EAF, Pahud BA, Llapur C, et al.; MATISSE Study Group. Bivalent prefusion F vaccine in pregnancy to prevent RSV illness in infants. N Engl J Med. 2023;388:1451–64. DOIPubMedGoogle Scholar
- Gilman MS, Castellanos CA, Chen M, Ngwuta JO, Goodwin E, Moin SM, et al. Rapid profiling of RSV antibody repertoires from the memory B cells of naturally infected adult donors. Sci Immunol. 2016;1:eaaj1879.
Figure
Table
Cite This ArticleOriginal Publication Date: September 13, 2023
1These first authors contributed equally to this article.
Table of Contents – Volume 29, Number 11—November 2023
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Efrem S. Lim, Arizona State University, PO Box 876101, Tempe, AZ 85287, USA
Top