Sentinel Surveillance System Implementation and Evaluation for SARS-CoV-2 Genomic Data, Washington, USA, 2020–2021
Hanna N. Oltean

, Krisandra J. Allen, Lauren Frisbie, Stephanie M. Lunn, Laura Marcela Torres, Lillian Manahan, Ian Painter, Denny Russell, Avi Singh, JohnAric MoonDance Peterson, Kristin Grant, Cara Peter, Rebecca Cao, Katelynn Garcia, Drew Mackellar, Lisa Jones, Holly Halstead, Hannah Gray, Geoff Melly, Deborah Nickerson, Lea Starita, Chris Frazar, Alexander L. Greninger, Pavitra Roychoudhury, Patrick C. Mathias, Michael H. Kalnoski, Chao-Nan Ting, Marisa Lykken, Tana Rice, Daniel Gonzalez-Robles, David Bina, Kelly Johnson, Carmen L. Wiley, Shaun C. Magnuson, Christopher M. Parsons, Eugene D. Chapman, C. Alexander Valencia, Ryan R. Fortna, Gregory Wolgamot, James P. Hughes, Janet G. Baseman, Trevor Bedford, and Scott Lindquist
Author affiliations: Washington State Department of Health, Shoreline, Washington, USA (H.N. Oltean, K.J. Allen, L. Frisbie, S.M. Lunn, L.M. Torres, L. Manahan, I. Painter, D. Russell, A. Singh, J.M. Peterson, K. Grant, C. Peter, R. Cao, K. Garcia, D. Mackellar, L. Jones, H. Halstead, H. Gray, G. Melly, S. Lindquist); University of Washington, Seattle, Washington (H.N. Oltean, D. Nickerson, L. Starita, C. Frazar, A.L. Greninger, P. Roychoudhury, P.C. Mathias, J.P. Hughes, J.G. Baseman); Northwest Genome Center, Seattle (D. Nickerson, L. Starita, C. Frazar); Atlas Genomics, Seattle (M.H. Kalnoski, C.-N. Ting); Confluence Health, Wenatchee, Washington (M. Lykken, T. Rice, D. Gonzalez-Robles); Incyte Diagnostics, Spokane Valley, Washington (D. Bina, K. Johnson, C.L. Wiley); Interpath Laboratory, Boise, Idaho, USA (S.C. Magnuson, C.M. Parsons, E.D. Chapman, A. Valencia); Northwest Laboratory, Bellingham, Washington (R.R. Fortna, G. Wolgamot); Fred Hutchinson Cancer Research Center, Seattle (T. Bedford)
Main Article
Figure 3
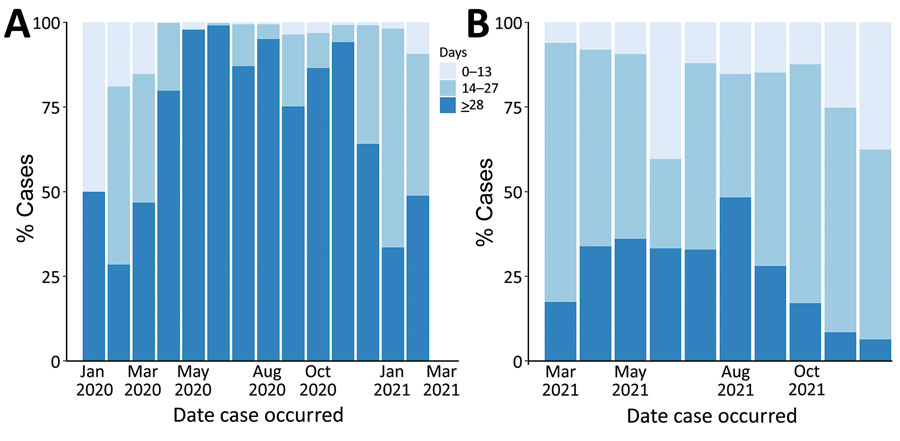
Figure 3. Timeliness of sequence data availability in study of sentinel surveillance system implementation and evaluation for SARS-CoV-2 genomic data, Washington, USA, 2020–2021. Graph shows percentages of COVID-19 cases with sequenced data uploaded to the GISAID database (https://www.gisaid.org) within 0–13, 14–27, and >28 days after specimen collection. A) Presentinel surveillance (specimens sequenced before March 1, 2021). B) Sentinel surveillance (specimens sequenced on or after March 1, 2021, through the sentinel surveillance program).
Main Article
Page created: December 13, 2022
Page updated: January 21, 2023
Page reviewed: January 21, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
