Volume 29, Number 3—March 2023
Dispatch
Mycobacterium leprae in Armadillo Tissues from Museum Collections, United States
Figure 2
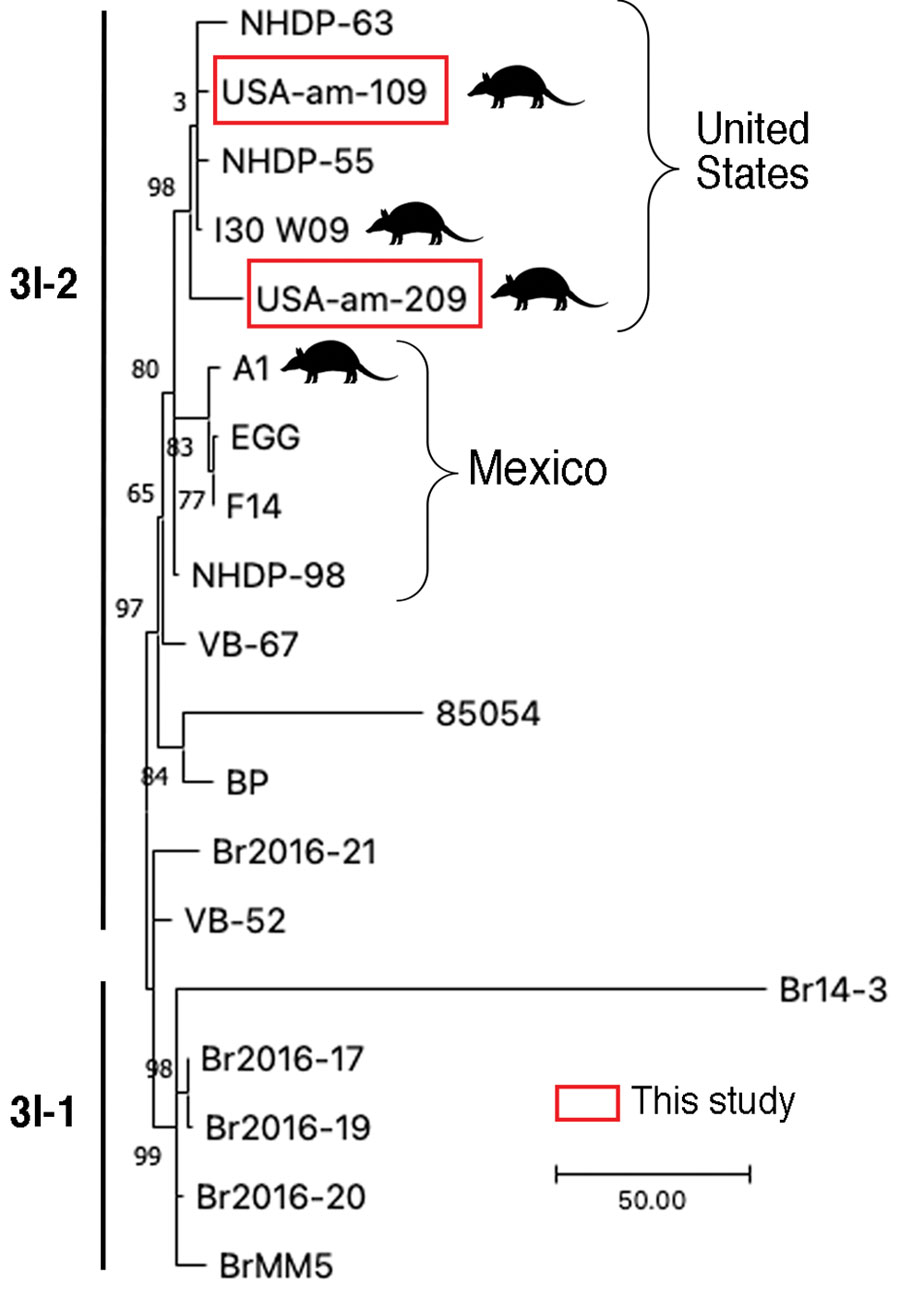
Figure 2. Comparative genomics of the Mycobacterium leprae sequenced this study from armadillo tissues from US museums and those from humans and armadillos from the United States and Mexico. Samples subjected to whole-genome sequencing, USA-am-109 and USA-am-209, clustered among genomes from humans and armadillos from the United States (branch 3I). The tree represents a zoom into the M. leprae genotypes 3I-1 and 3I-2 from a maximum-parsimony tree of 302 M. leprae genomes rooted with M. lepromatosis as outgroup. The tree was built in MEGA version 11 software (https://www.megasoftware.net). Support values were obtained by bootstrapping 500 replicates. Scale bar indicates number of nucleotide substitutions.
Page created: January 12, 2023
Page updated: February 20, 2023
Page reviewed: February 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.