Volume 29, Number 3—March 2023
Dispatch
Mycobacterium leprae in Armadillo Tissues from Museum Collections, United States
Cite This Article
Citation for Media
Abstract
We examined armadillos from museum collections in the United States using molecular assays to detect leprosy-causing bacilli. We found Mycobacterium leprae bacilli in samples from the United States, Bolivia, and Paraguay; prevalence was 14.8% in nine-banded armadillos. US isolates belonged to subtype 3I-2, suggesting long-term circulation of this genotype.
Hansen disease (leprosy) is an ancient pathology caused by 2 slow-growing intracellular bacilli, Mycobacterium leprae and M. lepromatosis (1). Both pathogens have the ability to damage peripheral nerves of hosts, producing a broad spectrum of clinical outcomes. Routes of disease transmission have been hypothesized for >100 years but are still actively debated (2); traditionally, human-to-human transmission has been considered the dominant route of infection. Evidence incriminates M. leprae as a zoonotic pathogen; the nine-banded armadillo (Dasypus novemcinctus) is its main wildlife reservoir in the southern United States (2). M. leprae also has been found in D. novemcinctus armadillos outside the United States (e.g., in Brazil), in the six-banded armadillo (Euphractus sexcinctus), and in nonhuman primates including chimpanzees, macaques, and sooty mangabeys (2). In addition, M. leprae and M. lepromatosis have been reported in red squirrels (Sciurus vulgaris) in the British Isles (3). Those data strongly suggest broad zoonotic transmission dynamics for both bacilli.
Natural history collections represent a neglected resource for biomedical research despite their known utility (4). We examined armadillo (family Dasypodidae) tissues deposited in museum collections across the United States to identify M. leprae and M. lepromatosis across space and time. We report presence of M. leprae in armadillo tissue samples from endemic and nonendemic areas of the Americas, suggesting that public health policy should contemplate zoonotic leprosy transmission routes carefully.
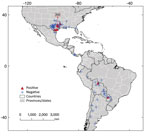
We assembled a database of museum armadillo tissue samples using the biodiversity information portals VertNet (http://portal.vertnet.org) and Arctos (https://arctos.database.museum/home.cfm), queried during December 2018–April 2019. Ten US museums included armadillo samples in their datasets. The samples were collected during 1974–2017 (Appendix 1 Figure 4) from 8 countries across the Americas; 68.6% (n = 109) came from the United States (Table 1; Figure 1). Each museum contributed ≈1 mm3 of armadillo tissue (Appendix 1; Appendix 2). The 159 samples processed corresponded to 10 armadillo species; D. novemcinctus, the nine-banded armadillo, was the most common (n = 122 [76.7%]). Most samples were liver tissues (n = 66 [41.5%]), followed by muscle (n = 37 [23.3%]) and spleen (n = 31 [19.5%]) (Table 1). The specimens were frozen or preserved in 10% dimethyl sulfoxide or 70%, 90%, or 95% ethanol; most were either frozen (n = 77 [48.4%]) or in 95% ethanol (n = 55 [34.6%]) (Table 1).
We processed tissues using an in-house DNA extraction method based on magnetic beads (Appendix 1). We applied standardized PCR protocols using specific primers to detect M. leprae and M. lepromatosis (5,6). For M. leprae, we implemented typification and subtypification as described previously (7). We performed quantitative real-time PCR (qPCR) on all samples for which genotyping was successful as a proxy of M. leprae DNA quantity with cycle threshold (Ct) <26 as a threshold for whole-genome sequencing. We multiplexed and sequenced libraries on an Illumina NextSeq 500 instrument (https://www.illumina.com) (Appendix 1).
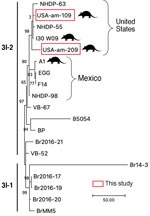
We found M. leprae in 18/159 (11.3%) samples. All positives were in D. novemcinctus armadillos, for prevalence in that species of 14.8% (n = 18/122). We detected positive results mainly in muscle tissue (n = 13/18 [72.2%]) and in 95% ethanol–preserved specimens (n = 13/18 [72.2%]) (Tables 1, 2). M. lepromatosis was not detected in the tissues examined. PCR subtyping was successful in 5/18 (27.8%) positive samples; 4 belonged to subtype 3I, as expected for armadillos from Texas, USA (8) (Table 2). The remaining sample was characterized only to type (3 or 4), because we found low amounts of M. leprae DNA (Table 2). After RLEP qPCR, 2 samples had a Ct<26 (i.e., 109 and 209) and were suitable for whole-genome sequencing. The genomes of M. leprae National Center for Biotechnology Information BioSample no. SAMN31421191 (https://www.ncbi.nlm.nih.gov/biosample) had coverage of 18.2× and of BioSample SAMN31421192, 4.9× (Appendix 1). Phylogenetic analysis showed that both M. leprae strains belonged to genotype 3I-2 (8,9). The 2 M. leprae genomes clustered specifically with other isolates previously identified in armadillos (i.e., I-30) and humans (i.e., NHDP-55 and NHDP-63) from the United States (Figure 2). Isolate 109 harbored 3 specific single-nucleotide polymorphisms, including 1 missense mutation in argD (i.e., C1691069T; Arg61Cys), encoding a probable acetylornithine aminotransferase. Sequence data are available from the National Center for Biotechnology Information Sequence Read Archive under accession no. PRJNA893376.
We identified M. leprae in D. novemcinctus armadillos only; prevalence was 14.8%. Positive samples were mainly detected from muscle and from ethanol-preserved specimens (Table 1). Infected armadillos were found in the United States, Paraguay, and Bolivia. M. leprae has not been reported in other wildlife in Paraguay or Bolivia. In our study, tissues from Paraguay were collected in 1996 and from Bolivia in 1993 (Table 2). Hansen disease is prevalent in humans in both countries (10); presence of infected armadillos should prompt research to explore their role as a potential zoonotic source of leprosy (2). In Bolivia, a previous survey of D. novemcinctus and T. matacus armadillos conducted during 1999–2001 found 0 positive animals (2). We found 7 M. leprae–negative armadillo tissues in the United States: 1 from Florida in 1974 and 6 from Texas collected during 1982–1990 (Appendix 2). No evidence for M. leprae was reported in Florida before 2009 (8). In Texas, although immunologic detection studies suggested the presence of M. leprae in armadillos before the 2000s, evidence was restricted to 1 area (2). Thus, our molecular identification of M. leprae in Texas armadillos from 1996, 1999, and 2000 are novel records (Table 2; Figure 1).
M. lepromatosis has been reported in multiple countries of the Americas, including the United States, Mexico, and Colombia, but as of 2022, only in humans (11,12). Although this species has been detected in Sciurus vulgaris squirrels in the British Isles, broader surveillance in rodents across Europe and Mexico identified 0 positive samples (13). From our dataset we obtained only negative results. M. lepromatosis is seldom screened as a Hansen disease–causing pathogen because of lack of awareness, which has impeded understanding of its incidence. Thus, in countries endemic for Hansen disease, M. lepromatosis should be also screened systematically in humans and potential animal reservoirs.
We were able to identify M. leprae subtypes in 4 armadillos (Table 2) and to sequence 2 entire genomes. Those 2 strains clustered with armadillo and human isolates from the United States, all belonging to subtype 3I-2, on branch 3 of the genetic tree (Figure 2). Of interest, our isolates differed by several nonsynonymous sites from those isolated previously. Our findings corroborate that several strains of M. leprae are circulating in armadillo populations in the southern United States (8,9,14). As predicted (15), our data also confirm that the strains circulating in armadillos today are close to those infecting animals >30 years ago, highlighting the promise of using preserved animal tissues to study epizootic dynamics of leprosy and other diseases.
Information on pathogen biodiversity in wildlife is much needed. We suggest that specimens in natural museums can play a role in infectious disease monitoring; our study relied on the global museum initiative and the large digital repositories of relevant specimen data in the United States. Protocols for using museum repositories for infectious disease research are still in development (4); parameters to optimal pathogen identification should be explored for M. leprae and other pathogens. We recognize that no single best way to study the diversity of pathogens exists; any approach should consider the specific nuances of each zoonotic system.
Dr. Romero-Alvarez is an MD and PhD candidate at the Biodiversity Institute and the Department of Ecology & Evolutionary Biology at the University of Kansas and is affiliated with the Universidad de las Américas, Quito, Ecuador. His research is focused on the eco-epidemiology and geographic distribution of infectious diseases.
Acknowledgments
The reagent genomic DNA from Mycobacterium leprae, strain Thai-53, NR-19352 was obtained through BEI Resources, US National Institute of Allergy and Infectious Diseases, National Institutes of Health. Genomic DNA for Mycobacterium lepromatosis was provided by Ramanuj Lahiri (National Hansen’s Disease Program, Baton Rouge, Louisiana, USA).
C.A. and M.J. are supported by the Fondation Raoul Follereau, the Heiser Program of the New York Community Trust for Research in Leprosy (grant no. P21-000127), the European Union’s Horizon 2020 Research and Innovation Program (C.A. by Marie Sklodowska-Curie grant no. 845479).
References
- Singh P, Benjak A, Schuenemann VJ, Herbig A, Avanzi C, Busso P, et al. Insight into the evolution and origin of leprosy bacilli from the genome sequence of Mycobacterium lepromatosis. Proc Natl Acad Sci U S A. 2015;112:4459–64. DOIPubMedGoogle Scholar
- Ploemacher T, Faber WR, Menke H, Rutten V, Pieters T. Reservoirs and transmission routes of leprosy; A systematic review. PLoS Negl Trop Dis. 2020;14:
e0008276 . DOIPubMedGoogle Scholar - Avanzi C, Del-Pozo J, Benjak A, Stevenson K, Simpson VR, Busso P, et al. Red squirrels in the British Isles are infected with leprosy bacilli. Science. 2016;354:744–7. DOIPubMedGoogle Scholar
- Colella JP, Bates J, Burneo SF, Camacho MA, Carrion Bonilla C, Constable I, et al. Leveraging natural history biorepositories as a global, decentralized, pathogen surveillance network. PLoS Pathog. 2021;17:
e1009583 . DOIPubMedGoogle Scholar - Sharma R, Singh P, McCoy RC, Lenz SM, Donovan K, Ochoa MT, et al. Isolation of Mycobacterium lepromatosis and development of molecular diagnostic assays to distinguish Mycobacterium leprae and M. lepromatosis. Clin Infect Dis. 2020;71:e262–9. DOIPubMedGoogle Scholar
- Tió-Coma M, Wijnands T, Pierneef L, Schilling AK, Alam K, Roy JC, et al. Detection of Mycobacterium leprae DNA in soil: multiple needles in the haystack. Sci Rep. 2019;9:3165. DOIPubMedGoogle Scholar
- Monot M, Honoré N, Garnier T, Zidane N, Sherafi D, Paniz-Mondolfi A, et al. Comparative genomic and phylogeographic analysis of Mycobacterium leprae. Nat Genet. 2009;41:1282–9. DOIPubMedGoogle Scholar
- Sharma R, Singh P, Loughry WJ, Lockhart JM, Inman WB, Duthie MS, et al. Zoonotic leprosy in the southeastern United States. Emerg Infect Dis. 2015;21:2127–34. DOIPubMedGoogle Scholar
- Vera-Cabrera L, Ramos-Cavazos CJ, Youssef NA, Pearce CM, Molina-Torres CA, Avalos-Ramirez R, et al. Mycobacterium leprae infection in a wild nine-banded armadillo, Nuevo León, Mexico. Emerg Infect Dis. 2022;28:747–9. DOIPubMedGoogle Scholar
- Schaub R, Avanzi C, Singh P, Paniz-Mondolfi A, Cardona-Castro N, Legua P, et al. Leprosy transmission in Amazonian countries: current status and future trends. Curr Trop Med Rep. 2020;7:79–91. DOIGoogle Scholar
- Cardona-Castro N, Escobar-Builes MV, Serrano-Coll H, Adams LB, Lahiri R. Mycobacterium lepromatosis as cause of leprosy, Colombia. Emerg Infect Dis. 2022;28:1067–8. DOIPubMedGoogle Scholar
- Deps P, Collin SM. Mycobacterium lepromatosis as a second agent of Hansen’s disease. Front Microbiol. 2021;12:
698588 . DOIPubMedGoogle Scholar - Schilling AK, Avanzi C, Ulrich RG, Busso P, Pisanu B, Ferrari N, et al. British red squirrels remain the only known wild rodent host for leprosy bacilli. Front Vet Sci. 2019;6:8. DOIPubMedGoogle Scholar
- Truman RW, Andrews PK, Robbins NY, Adams LB, Krahenbuhl JL, Gillis TP. Enumeration of Mycobacterium leprae using real-time PCR. PLoS Negl Trop Dis. 2008;2:
e328 . DOIPubMedGoogle Scholar - Schuenemann VJ, Singh P, Mendum TA, Krause-Kyora B, Jäger G, Bos KI, et al. Genome-wide comparison of medieval and modern Mycobacterium leprae. Science. 2013;341:179–83. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: February 15, 2023
Table of Contents – Volume 29, Number 3—March 2023
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Daniel Romero-Alvarez, Biodiversity Institute and Department of Ecology & Evolutionary Biology, University of Kansas, 1345 Jayhawk Blvd, Lawrence, KS 66044, USA
Top