Volume 29, Number 6—June 2023
Dispatch
Replication of Novel Zoonotic-Like Influenza A(H3N8) Virus in Ex Vivo Human Bronchus and Lung
Cite This Article
Citation for Media
Abstract
Human infection with avian influenza A(H3N8) virus is uncommon but can lead to acute respiratory distress syndrome. In explant cultures of the human bronchus and lung, novel H3N8 virus showed limited replication efficiency in bronchial and lung tissue but had a higher replication than avian H3N8 virus in lung tissue.
Avian influenza viruses (AIVs) with reassortments between AIVs from domestic poultry and wild birds sporadically cross species barriers, leading to human infections. Viruses with internal genes of H9N2, hemagglutinin, and neuraminidase acquired from wild birds constitute the zoonotic H5N1, H7N9, and H10N8 viruses (1–3) and can lead to severe influenza.
In 2022, two human infections with novel influenza A(H3N8) viruses were reported in Henan and Hunan Province, China (4,5). The first case was identified in a 4-year-old boy with acute respiratory distress syndrome, and the second case occurred in a 5-year-old boy with mild disease. Phylogenetic analysis revealed that the novel H3N8 viruses were triple reassortments containing the Eurasian avian H3 gene of wild-bird origin, the North American avian N8 gene derived from the wild bird AIV, and G57 genotype H9N2 internal genes from AIVs found in poultry in China (6,7). H3N8 viruses that are genetically similar to the zoonotic H3N8 viruses reported in China (4,5) have been isolated in poultry markets in Hong Kong, China (8). Those novel avian H3N8 viruses are antigenically distant from contemporary human influenza A(H3N2) viruses, and little cross-reactive immunity to these chicken H3N8 viruses exists in the human population (8). We assessed the replication of the novel influenza A(H3N8) virus in human ex vivo bronchus and lung tissues (Appendix).
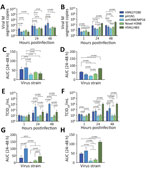
The viruses used in this study were H9N2/Y280, pH1N1, avH3N8/MP16, novel H3N8, and H5N1/483 (Appendix Table 1). The novel H3N8 virus was isolated from chickens and is genetically closely related to the virus causing zoonotic human disease in China (A/Henan/4-10CNIC/2022/H3N8) (8). Their hemagglutinin genes share a 99.1% similarity, and the neuraminidase genes share a 98.7% similarity. The avH3N8 virus was isolated from wild bird droppings in Mai Po, Hong Kong, and is genetically unrelated to the virus causing zoonotic disease in China. The novel H3N8 virus failed to propagate in Madin-Darby canine kidney (MDCK) cells but could be propagated in eggs and titrated in chicken embryo fibroblasts (DF-1), whereas the other strains could be propagated and titrated in MDCK cells. We performed titration in cells that support the replication of the influenza A viruses rather than in all DF-1 cells, because pH1N1 virus did not replicate in DF-1 cells. We investigated the virus replication kinetics by measuring viral matrix protein segment RNA in culture supernatants using real-time quantitative reverse transcription PCR and 50% tissue culture infectious dose (TCID50) assay for infectious virus titers (Figure 1).
In bronchial tissues, pH1N1 virus had a higher level of viral RNA than did avH3N8, novel H3N8, and H5N1, whereas levels of viral RNA of H9N2 virus were higher than those of avH3N8 and H5N1 virus (Figure 1, panels A, C). The viral RNA levels of avH3N8, novel H3N8, and H5N1 virus were similar. The viral RNA level of H5N1 virus was the highest among all the tested strains in human lung tissues, followed by H9N2 (Figure 1, panels B, D). Viral RNA levels of novel H3N8 and pH1N1 viruses were higher than those of avH3N8 virus. Measurement of viral RNA using quantitative reverse transcription PCR is sensitive, but it cannot distinguish defective viral particles from infectious ones. Therefore, we performed the TCID50 assay to monitor the infectious viral titers.
As expected, replication of pH1N1 virus was the highest among all tested strains in human bronchial tissues, in both viral titers and area under the curve values (Figure 1, panels E, G). The titers of H5N1 virus were similar to those of H9N2 virus, whereas H5N1 virus had higher replication competence than did avH3N8 and novel H3N8 viruses. We observed a discrepant trend between viral RNA copies and infectious titers for H9N2. Viral RNA levels of H9N2 were similar to those of pH1N1 (Figure 1, panels A, C), but the infectious titers of H9N2 virus were significantly lower than that of pH1N1 in bronchus (Figure 1, panels E, G).
In lung tissue, H5N1 virus had the highest replication of all strains tested (Figure 1, panels F, H). Similar to pH1N1 virus, H9N2 had higher titers than the 2 H3N8 viruses in lung tissues. avH3N8 had the lowest titer measured by TCID50. The novel H3N8 virus replicated poorly in mammalian MDCK cells but replicated efficiently in DF-1 avian cells (Appendix Figure). Those findings imply that the novel H3N8 virus has not yet adapted to mammal hosts, which was confirmed by limited replication in human bronchial and lung tissues (Figure 1, panels E, F).
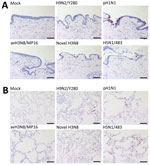
We fixed infected tissues at 48 hours postinfection and stained them for influenza A nucleoprotein immunohistochemistry (Appendix). Consistent with TCID50 findings, bronchial tissues infected with pH1N1 had the most extensive distribution of viral antigen, whereas we observed moderate levels of viral antigen staining in tissues infected with novel H3N8, H9N2, and H5N1 virus. No viral antigen staining was observed in tissues infected with avH3N8 (Figure 2, panel A). In the lung, H5N1 virus infection demonstrated the most extensive viral antigen staining, followed by infection with pH1N1, novel H3N8, and avH3N8 virus, which demonstrated the least extensive staining (Figure 2, panel B).
The discrepancy between the viral load in RNA copies and TCID50 titers of H9N2 and avH3N8 infection suggests that infection with those viruses might produce high levels of defective particles that cannot be detected by TCID50 assay. Immunohistochemistry staining of viral antigen serves as alternative evidence of virus replication in human tissues. The staining correlates more with TCID50 results than with viral RNA analysis for all the viruses.
Amino acid comparisons of the novel H3N8 and avH3N8 viruses demonstrated that they shared the same stalk length in the NA gene but did not have the G228S mutation that enhances binding to mammalian receptors (Appendix Table 2). The internal genes of the novel H3N8 virus were reassorted from H9N2 virus, whereas the internal genes of the avH3N8 came from H3N8, H6N1, H6N2, H3N8, H1N1, and H7N1 (Table). Neither virus had the E627K mutation in polymerase basic 2 that confers mammal adaptation, virulence, and transmissibility. The novel H3N8 virus had the A588V mutation in polymerase basic 2 that promotes mammal adaptation, but avian H3N8 virus did not have this mutation. This difference might contribute to higher replication of the novel H3N8 virus in human lung tissue. The S31N mutation found in the matrix protein 2 of the novel H3N8 virus provided adamantane resistance.
Although zoonotic H3N8 viruses have a dual receptor-binding affinity of α-2,3 and α-2,6 receptors (7), our findings show that this factor does not confer an advantage for replication in human bronchial tissue. Our findings demonstrated inefficient replication of the novel H3N8 virus in human bronchial tissues, which implies limited efficiency to transmit among humans. This finding is in line with a recent serologic surveillance study in which no poultry workers were positive for antibodies for the novel H3N8 virus (7), and only 2 human cases have been documented since April 2022 (4,5). The moderate replication ability of the novel H3N8 virus in human lung tissue suggests that the virus causes less severe disease than H5N1 virus.
In summary, our findings suggest that the zoonotic-like avian H3N8 virus has limited efficiency for human-to-human transmission and, at present, is unlikely to cause severe disease in humans. However, the limited cross-reactive immunity against this novel H3N8 virus in the human population (8) and the emergence of novel H3N8 viruses by continuous reassortment between AIVs in wild birds and poultry demonstrate that the zoonotic and pandemic potential of avian H3N8 viruses should be closely monitored.
Dr. Hui is an assistant professor in the School of Public Health at the University of Hong Kong. Her research interests include risk assessment, understanding the pathogenesis of emerging respiratory viruses, and the development of therapeutic options for severe influenza diseases and coronavirus infections.
Acknowledgments
We thank Jenny Chan, Rachel Ching, and Kevin Fung for technical support.
This work was supported by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (contract no. 75N93021C00016) and the Theme-Based Research Scheme (Ref: T11-712/19-N) under University Grants Committee of Hong Kong Special Administrative Region.
References
- Guan Y, Shortridge KF, Krauss S, Webster RG. Molecular characterization of H9N2 influenza viruses: were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc Natl Acad Sci U S A. 1999;96:9363–7.
- Lam TT, Zhou B, Wang J, Chai Y, Shen Y, Chen X, et al. Dissemination, divergence and establishment of H7N9 influenza viruses in China. Nature. 2015;522:102–5.
- Ma C, Lam TT, Chai Y, Wang J, Fan X, Hong W, et al. Emergence and evolution of H10 subtype influenza viruses in poultry in China. J Virol. 2015;89:3534–41.
- Cheng D, Dong Y, Wen S, Shi C. A child with acute respiratory distress syndrome caused by avian influenza H3N8 virus. J Infect. 2022;85:174–211.
- Tan X, Yan X, Liu Y, Wu Y, Liu JY, Mu M, et al. A case of human infection by H3N8 influenza virus. Emerg Microbes Infect. 2022;11:2214–7.
- Li Y, Li P, Xi J, Yang J, Wu H, Zhang Y, et al. Wild bird-origin H3N8 avian influenza virus exhibit well adaptation in mammalian host. J Infect. 2022;84:579–613.
- Yang R, Sun H, Gao F, Luo K, Huang Z, Tong Q, et al. Human infection of avian influenza A H3N8 virus and the viral origins: a descriptive study. Lancet Microbe. 2022;3:e824–34.
- Sit THC, Sun W, Tse ACN, Brackman CJ, Cheng SMS, Tang AWY, et al. Novel zoonotic avian influenza A(H3N8) virus in chicken, Hong Kong, China. Emerg Infect Dis. 2022;28:2009–15.
Figures
Table
Cite This ArticleOriginal Publication Date: April 24, 2023
Table of Contents – Volume 29, Number 6—June 2023
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Michael C.W. Chan, School of Public Health, Li Ka Shing Faculty of Medicine, The University of Hong Kong, Pok Fu Lam, Hong Kong, China
Top