Early SARS-CoV-2 Reinfections Involving the Same or Different Genomic Lineages, Spain
Cristina Rodríguez-Grande, Agustín Estévez, Rosalía Palomino-Cabrera, Andrea Molero-Salinas, Daniel Peñas-Utrilla, Marta Herranz, Amadeo Sanz-Pérez, Luis Alcalá, Cristina Veintimilla, Pilar Catalán, Carolina Martínez-Laperche, Roberto Alonso, Patricia Muñoz, Laura Pérez-Lago
1
, Darío García de Viedma
1
, and
on behalf of the Gregorio Marañón Microbiology-ID COVID 19 Study Group2
Author affiliations: Gregorio Marañón General University Hospital, Madrid, Spain (C. Rodríguez-Grande, A. Estévez, R. Palomino-Cabrera, A. Molero-Salinas, D. Peñas-Utrilla, M. Herranz, A. Sanz-Pérez, L. Alcalá, C. Veintimilla, P. Catalán, C. Martínez-Laperche, R. Alonso, P. Muñoz, L. Pérez-Lago, D. García de Viedma); Instituto de Investigación Sanitaria Gregorio Marañón (IiSGM), Madrid (C. Rodríguez-Grande, A. Estévez, R. Palomino-Cabrera, A. Molero-Salinas, D. Peñas-Utrilla, A. Sanz-Pérez, L. Pérez-Lago, D. García de Viedma); Universidad Complutense, Madrid (R. Alonso, P. Muñoz); Centro de Investigación Biomédica en Red (CIBER) de Enfermedades Respiratorias-CIBERES, Madrid (R. Alonso, P. Muñoz, D. García de Viedma)
Main Article
Figure
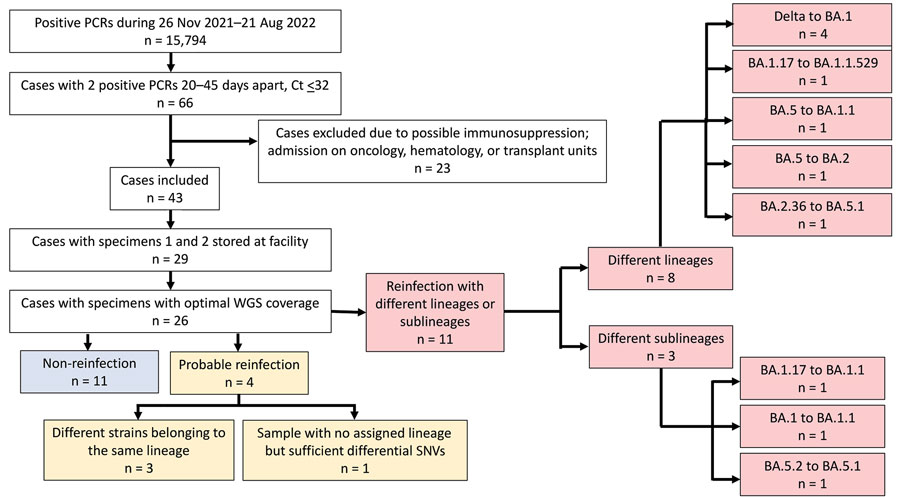
Figure. Flowchart of case selection in a study of early SARS-CoV-2 reinfection involving the same or different genomic lineages, Spain. PCR-positive cases were diagnosed by our tertiary hospital, which covers 650,000 inhabitants in the population of Madrid. Among 26 cases with optimal coverage for WGS, 11 were reinfections (red boxes), 4 of which were non-Omicron to Omicron lineage reinfections. Probable reinfection cases (yellow boxes; patients 23–26) showed enough unique SNV differences between the sequences from their sequential specimens to be suspect of reinfection (Appendix Table 3). Ct, cycle threshold; SNV, single-nucleotide variants; WGS, whole-genome sequencing.
Main Article
Page created: April 17, 2023
Page updated: May 17, 2023
Page reviewed: May 17, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
