Population Structure and Antimicrobial Resistance in Campylobacter jejuni and C. coli Isolated from Humans with Diarrhea and from Poultry, East Africa
Nigel P. French

, Kate M. Thomas, Nelson B. Amani, Jackie Benschop, Godfrey M. Bigogo, Sarah Cleaveland, Ahmed Fayaz, Ephrasia A. Hugho, Esron D. Karimuribo, Elizabeth Kasagama, Ruth Maganga, Matayo L. Melubo, Anne C. Midwinter, Blandina T. Mmbaga, Victor V. Mosha, Fadhili I. Mshana, Peninah Munyua, John B. Ochieng, Lynn Rogers, Emmanuel Sindiyo, Emanuel S. Swai, Jennifer R. Verani, Marc-Alain Widdowson, David A. Wilkinson, Rudovick R. Kazwala, John A. Crump
1, and Ruth N. Zadoks
1
Author affiliations: Massey University, Palmerston North, New Zealand (N.P. French, J. Benschop, A. Fayaz, A.C. Midwinter, L. Rogers, D.A. Wilkinson); Ministry for Primary Industries, Wellington, New Zealand (K.M. Thomas); Kilimanjaro Clinical Research Institute, Kilimanjaro Christian Medical Centre, Moshi, Tanzania (N.B. Amani, E.A. Hugho, E. Kasagama, M.L. Melubo, B.T. Mmbaga, V.V. Mosha, F.I. Mshana); Centre for Global Health Research, Kenya Medical Research Institute, Kisumu, Kenya (G.M. Bigogo, J.B. Ochieng); University of Glasgow, Glasgow, Scotland, UK (S. Cleaveland, R. Maganga, R.N. Zadoks); Sokoine University of Agriculture, Morogoro, Tanzania (E.D. Karimuribo, R.R. Kazwala); Kilimanjaro Christian Medical University College, Moshi (B.T. Mmbaga); US Centers for Disease Control and Prevention, Nairobi, Kenya (P. Munyua, J.R. Verani, M.-A. Widdowson); Nelson Mandela African Institution of Science and Technology, Arusha, Tanzania (E. Sindiyo); Ministry of Livestock and Fisheries, Dodoma, Tanzania (E.S. Swai); University of Otago, Dunedin, New Zealand (J.A. Crump); University of Sydney, Sydney, New South Wales, Australia (R.N. Zadoks)
Main Article
Figure 5
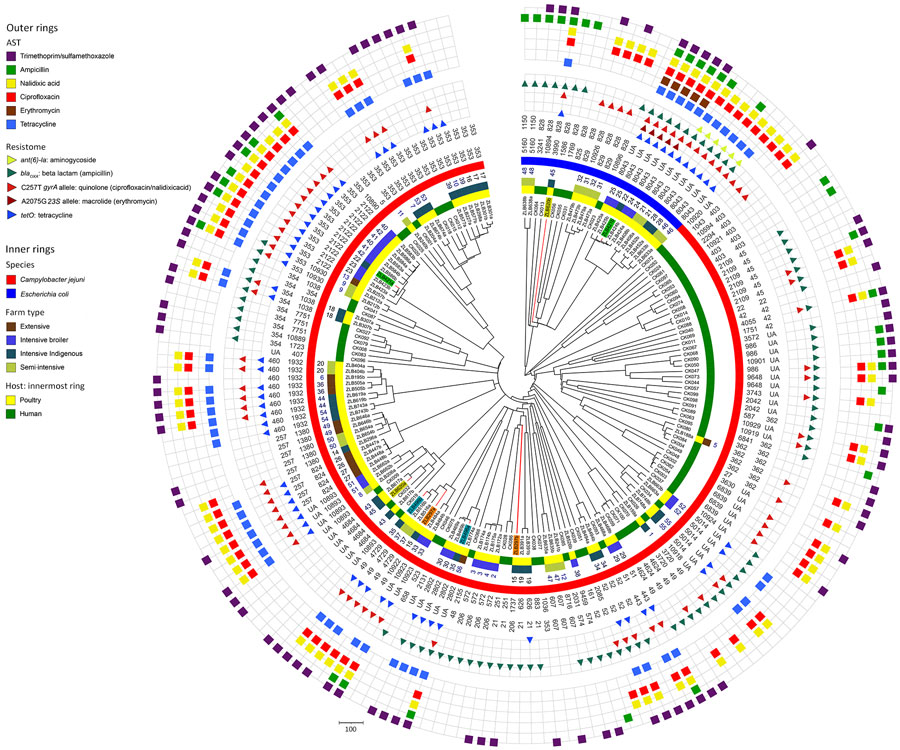
Figure 5. Circular dendrogram showing, from outer to inner rings, AST in study of population structure and antimicrobial resistance in Campylobacter jejuni and C. coli isolated from humans with diarrhea and from poultry, East Africa, 2006–2017. Colored blocks indicate resistance (all isolates were susceptible to gentamicin so this ring is not included), resistome, clonal complex (CC), sequence type (ST), Campylobacter species, poultry sample number, farm type, host and isolate ID for isolates from Kenya and Tanzania, 2006–2017 (human) or 2016–2017 (poultry). Isolates from the same poultry sample that belonged to a different ST are highlighted (samples 15, 22, 35, and 45) using colored isolate identification labels. The resistome indicates detection of resistance genes (encoding for resistance to some aminoglycosides, β-lactam antimicrobials and tetracyclines) and alleles (encoding for resistance for fluoroquinolones in the gyrase A gene, and macrolides in the 23S rRNA gene). Clustering of antimicrobial resistance phenotypes and the resistome with some CCs and STs is evident. For example, multidrug resistance is observed in C. jejuni ST2122 and C. coli ST8043 isolates. Scale bar indicates number of core-genome multilocus-sequence typing allele differences. AST, antimicrobial sensitivity.
Main Article
Page created: August 06, 2024
Page updated: September 24, 2024
Page reviewed: September 24, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
