Volume 30, Number 11—November 2024
Synopsis
Vibrio parahaemolyticus Foodborne Illness Associated with Oysters, Australia, 2021–2022
Cite This Article
Citation for Media
Abstract
The bacterium Vibrio parahaemolyticus is ubiquitous in tropical and temperate waters throughout the world and causes infections in humans resulting from water exposure and from ingestion of contaminated raw or undercooked seafood, such as oysters. We describe a nationwide outbreak of enteric infections caused by Vibrio parahaemolyticus in Australia during September 2021–January 2022. A total of 268 persons were linked with the outbreak, 97% of whom reported consuming Australia-grown oysters. Cases were reported from all states and territories of Australia. The outbreak comprised 2 distinct strains of V. parahaemolyticus, sequence types 417 and 50. We traced oysters with V. parahaemolyticus proliferation back to a common growing region within the state of South Australia. The outbreak prompted a national recall of oysters and subsequent improvements in postharvest processing of the shellfish.
Vibrio parahaemolyticus is a marine and estuarine bacterium that is ubiquitous in tropical and temperate waters worldwide (1). Infections, including wound infections, can occur in humans through exposure to water, and enteric infections are attributed most commonly to ingestion of raw or undercooked seafood. Invasive bloodstream infections and death rarely occur (2,3). Human infection is not associated with person-to-person spread or transmission through the fecal–oral route, and environmental presence of V. parahaemolyticus has not been linked to fecal contamination (4). The virulence of V. parahaemolyticus is associated with the presence of a thermostable direct hemolysin (coded by tdh gene) or thermostable related hemolysin (trh gene) (5).
Foodborne V. parahaemolyticus outbreaks have been reported across Asia, the United States, South America, Europe, and New Zealand, predominantly associated with consumption of filter-feeding bivalve shellfish, such as oysters and mussels (6–12). V. parahaemolyticus infections show seasonal patterns, with increases in warmer months, because V. parahaemolyticus will grow in seawater at temperatures >14°C–19°C (6). Climate change can increase the distribution and incidence of V. parahaemolyticus (6,13), and a rise in water temperature is the main environmental factor associated with growth of V. parahaemolyticus in oysters. Postharvest risk reduction strategies focus on rapid cooling and maintaining cold refrigeration of oysters throughout the supply chain to prevent bacterial growth, which occurs at air temperatures >10°C (14).
Until recently, foodborne outbreaks of V. parahaemolyticus were rare in Australia. Only 4 outbreaks were reported during 2002–2019, affecting a total of 24 persons (15). Two previously reported outbreaks were linked to consumption of oysters; 1 from oysters produced in Tasmania and 1 from oysters grown in South Australia. Only 29 locally acquired, sporadic foodborne cases of V. parahaemolyticus were reported in Australia in 2016–2020; 22 of the infected persons reporting oyster consumption (76%) (15). V. parahaemolyticus infections might be underreported in Australia because pathology laboratories rarely include it in routine fecal testing procedures and the infection is a notifiable condition in only 4 of the 8 states and territories of Australia.
In September 2021, at the end of the winter season in the Southern Hemisphere, health officials identified an increase in locally acquired V. parahaemolyticus cases in South Australia, and a similar trend was later noted in other jurisdictions of Australia. In November 2021, through the OzFoodNet network, a multijurisdictional outbreak investigation commenced to coordinate the public health response. OzFoodNet is a network of epidemiologists across Australia who are responsible for undertaking surveillance and outbreak investigations of foodborne disease (16). Investigators worked closely with jurisdictional public health laboratories and with the Australia food regulatory authorities who implement control measures.
V. parahaemolyticus infection is a notifiable disease under legislation in the Australia jurisdictions of the Northern Territory, South Australia, Tasmania, and Western Australia, where laboratories are required to report cases to their respective health departments. Public health authorities contacted diagnostic laboratories in the remaining Australia jurisdictions to request reported detections of V. parahaemolyticus in fecal specimens (under the auspices of an OzFoodNet multijurisdictional outbreak investigation).
Epidemiologic Investigation
We defined an outbreak case as illness in any person with a fecal specimen testing positive for V. parahaemolyticus during September 7, 2021–February 18, 2022. We conducted a descriptive case series investigation, which entailed telephone interviews of case-patient using a standardized questionnaire to obtain demographic information (age, sex, jurisdiction of residence), onset of illness, symptoms, medications, risk factors, and consumption of seafood during the exposure period (defined as 7 days before onset).
We classified cases as confirmed outbreak cases if single-nucleotide polymorphism (SNP) cluster analysis was performed on sequence type (ST) 417 or ST50 V. parahaemolyticus isolates on AusTrakka (a national genomic surveillance platform) and determined to be highly related within each ST. We considered outbreak cases as probable if they were typed as ST417 or ST50 without further phylogenetic analysis on AusTrakka and as possible if isolates were unable to be further typed (no ST). Cases were excluded if case-patients had traveled overseas in the 7 days before onset, if another ST was identified, or if the sequences did not cluster by phylogenetic analysis on AusTrakka. We used a broad case definition to include both STs in the outbreak investigation to describe the overall increase in locally acquired V. parahaemolyticus cases.
We entered case data into REDCap v10.3.4 (Vanderbilt University, https://projectredcap.org). We calculated proportions, medians, and ranges by using Stata BE v17 (Stata, https://www.stata.com). Where data were missing, we calculated a proportion with known responses only.
Environmental Investigations
Jurisdictional food regulatory authorities conducted traceback investigations for oyster exposures. Traceback activity revealed harvest area and dates, and investigators then sought information relative to temperature control. Food regulatory personnel collected oyster samples from case households and retail premises. The South Australian Shellfish Quality Assurance Program collected oyster samples direct from growers. Technicians processed and tested oyster samples in approved laboratories across jurisdictions to determine the presence of V. parahaemolyticus according to the Australian standards for food microbiology examination for specific organisms. Methods involved grinding 25 g of oysters with alkaline peptone water before overnight incubation at 36°C ± 2°C. Laboratory technicians isolated V. parahaemolyticus from the ground samples on thiosulfate–citrate–bile salts–sucrose agar at 36°C ± 2°C. They collected positive green-colonies and sent the samples to public health laboratories for whole-genome sequencing (WGS).
Genomic Sequencing and Analysis of V. parahaemolyticus
Public health laboratories in each jurisdiction sequenced V. parahaemolyticus isolates from cases and food samples for species confirmation and ST determination through multilocus sequence typing (https://github.com/tseemann/mlst) (17). Technicians performed WGS by using Illumina platforms (MiSeq and NextSeq 500/550; Illumina, https://illumina.com) with paired-end reads. Public health laboratories shared the raw sequencing reads of most isolates to AusTrakka (18). The national analysis team performed quality filtering, virulence gene detection, and phylogenetic analyses by using Snippy version 4.6.0 (https://github.com/tseemann/snippy) for cluster identification. Laboratory researchers determined genomic clusters for respective STs by using single-linkage clustering based on the SNP distance threshold of 5 SNPs, 10 SNPs, and 20 SNPs. The laboratories also performed pangenome analysis by using Roary version 3.13.0 (https://sanger-pathogens.github.io/Roary) (19) to include other publicly available V. parahaemolyticus genomes from PubMLST (https://pubmlst.org) and visualized a phylogenetic tree by using ggtree (https://guangchuangyu.github.io/software/ggtree). Genome sequences were uploaded to the National Center for Biotechnology Information’s Sequence Read Archive under the BioProject Accession nos. PRJNA1129299, PRJNA783474, PRJNA856407, and PRJNA1131944.
Epidemiologic Investigation
We investigated a total of 268 outbreak cases from all Australia jurisdictions:184 confirmed cases, 29 probable cases, and 55 possible cases (Figure 1). The outbreak occurred over a 5-month period, and the peak of cases occurred in mid-November 2021. Infections of ST50 were reported initially, followed by predominant reports of ST417 infections; subsequent reports then revealed a period of high overlap of the 2 STs. We noted the highest percentage of reported cases from residents of South Australia (28%, n = 76), followed by 2 jurisdictions where V. parahaemolyticus is not notifiable, Victoria (26%, n = 69) and Queensland (22%, n = 59) (Table 1). Some case-patients reported spending their entire incubation period in other jurisdictions, including a Tasmania resident exposed in South Australia and a South Australia resident exposed in Western Australia. More case-patients were male (57%) than female (43%). All jurisdictions with >5 outbreak cases included cases of both ST50 and ST417. The median age of case-patients was 52 years (range 1–90 years) (Table 2).
Of those with available information, 25% of case patients (51/206) sought treatment at hospital emergency departments and 13% (27/209) of case-patients were hospitalized; emergency department information was not reported for 3 case-patients. Of 24 cases with length of hospitalization recorded, the median stay was 2.5 days (range 1–7 days). There were no deaths. Of those who responded to symptom-specific questions, all 195 case-patients interviewed reported diarrhea, and 85% (165) reported abdominal pain. One case-patient had V. parahaemolyticus isolated from both a fecal specimen and blood culture. The median duration of illness for 131 cases with data available was 7 days (range 1–17 days); however, 40 cases were still unwell at the time of interview and were therefore not included in this calculation.
Of 206 case-patients interviewed, 199 (97%) reported consuming oysters, 189 (92%) reported consuming at least some of the oysters raw, and 25 (12%) reported consuming oysters for >1 meal in the week before onset. For case-patients who consumed oysters on only a single occasion (n = 131), the median incubation period was 1 day (range 4 hours to 7 days). The median number of oysters eaten per case-patient was 6 (range 1–31 oysters). Case-patients purchased oysters at a range of venues, including restaurants (n = 71), supermarkets or seafood stores (n = 23), farms (n = 17), oyster tours (n = 9), and takeaway venues (n = 8). Exposure to other seafood was common, including 41% (n = 84) who reported consuming fish (varied types) and 37% (n = 76) who consumed prawns in the 7 days before onset. For those who consumed fish or prawns, most reported food to have been cooked. Less than 20% of case patients reported consuming seafood other than oysters, fish, or prawns (Table 3). From 166 case interviews, 25% (42) of case-patients reported they had taken medication that reduced stomach acid (e.g., reflux or ulcer medications) in the month before onset.
Environmental Investigation
Traceback of oysters was complex because the supply chain could include farmers, processors (harvesters), brokers, wholesalers, retailers, and food services. Brokers and processors could receive stock from multiple growers on the same day and often from different growing regions. Processors could manage multiple suppliers on the same day, and opportunities for traceability were sometimes lost because records lacked details of where batches had been distributed. Sometimes, processors recorded shuck dates but not harvest dates on packaging. There was no single method of easily identifying unlabeled oysters once original traceability was misplaced by the processor. Traceback indicated oysters had been sourced from different growing regions in South Australia, including Smoky Bay, Streaky Bay, and Coffin Bay, with the largest proportion traced back to Coffin Bay. Within Coffin Bay, there were 32 accredited growers, and it was impossible to definitively link oysters to any single grower. In total, 173 oyster exposures were able to be traced back to Coffin Bay.
Of 117 oyster samples tested for V. parahaemolyticus, 14 tested positive (7 from South Australia, 3 from Queensland, 3 from Victoria, 1 from Western Australia). All positive oysters were ST417. Those V. parahaemolyticus–positive oyster samples were from various sources, including case-patient households, retail vendors, distributors, and direct-purchase farms. We traced the original source of all 14 positive oyster samples to Coffin Bay.
The Department of Primary Industries and Regions of South Australia (PIRSA) closed the Coffin Bay growing area on November 16, 2021, for harvest of oysters. A national recall of Coffin Bay raw pacific oysters occurred on November 19, 2021, conducted via Emergency Orders under the South Australian Food Act 2001. PIRSA also served compliance orders on accredited growers in Coffin Bay on November 18, 2021, specifying legislative requirements for growers to resume harvesting. Growers were required to implement a Vibrio control program and provide evidence that they had infrastructure available to maintain cold chain, could address food safety requirements, could verify monitoring and traceability, and could validate refrigeration capabilities. The Vibrio control program required growers to place oysters under active refrigeration within 7 hours of harvest, ensure oysters were at ≤10°C within 24 hours of harvest, ensure oysters were dispatched and transported at ≤10°C, and ensure enhanced traceability by including the harvest date, area, and aquaculture license number on invoices. Vibrio control programs were implemented in all oyster-growing areas across South Australia during this outbreak investigation. PIRSA also conducted microbiological sampling of oysters to clear growing zones in Coffin Bay before emergency orders were able to be lifted.
Genomic Epidemiology and Pathogenicity
Most outbreak cases (79%) could be classified as confirmed or probable cases, including 143 cases typed as ST417 and 70 cases typed as ST50. Cases of both STs occurred throughout the duration of the outbreak (Figure 1). All isolates from the 14 positive oyster samples were ST417 V. parahaemolyticus.
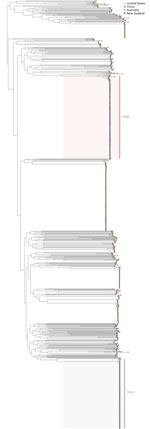
Phylogeny confirmed that ST417 and ST50 V. parahaemolyticus were not closely related (Figure 2). Because of the distinct nature of those strains of V. parahaemolyticus, we performed phylogenetic SNP clustering analyses on each ST individually. We grouped all clinical cases of ST417 and oyster samples from Australia submitted for national analysis in AusTrakka (n = 135) into a single cluster at a 5-SNP threshold. Clustering analysis of V. parahaemolyticus ST50 grouped 65 clinical cases at the 10-SNP threshold. Further analysis at the narrower 5-SNP threshold revealed 2 distinct but related clusters (n = 35 and n = 28; 2 were unclustered at 5 SNPs). We excluded 4 cases from the outbreak based on genomic analysis: 1 ST50 case unclustered at the 10-SNP threshold on phylogenetic analysis in AusTrakka and 3 cases that were different STs (2 ST1140 and 1 ST36).
Both STs of V. parahaemolyticus isolated from cases in this outbreak investigation harbored virulence genes required for pathogenicity. Specifically, all V. parahaemolyticus ST417 isolates harbored the virulence gene trh, and all ST50 harbored 2 virulence genes, tdh and trh.
This V. parahaemolyticus outbreak was caused by 2 STs and had a considerable effect on the population of Australia because of the nationwide distribution of oysters across mainland jurisdictions and cases occurring over a 5-month period. Recent investigations of other V. parahaemolyticus outbreaks have focused mostly on point source events, such as outbreaks on cruise ships (8,20,21). The communitywide outbreak in this report highlights the potential risks associated with consumption of raw oysters in Australia. Raw shellfish, particularly oysters, are known to be a common source of Vibrio foodborne illness, but recent trends observe increasing numbers of sporadic and outbreak cases, across an internationally wider span, somewhere cases had not been previously reported (8,11,22). V. parahaemolyticus has been isolated from other shellfish, including mussels, prawns, clams, and scallops during food surveillance studies, and has been identified as the cause of outbreaks in countries other than Australia (3,23–25).
The identification of 2 unrelated STs, ST417 and ST50, within this outbreak indicated the cause to be more relative to environmental factors influencing favorable growth conditions for V. parahaemolyticus across the oyster-growing region than to a single temperature-abuse error or single point source event. The appearance of those 2 strains could also be indicative of >1 outbreak occurring at the same time, with common contributing factors. However, multiple strains or types of a pathogen can cause discrete outbreaks and require a common public health investigation and response (26,27). The epidemiologic evidence in this investigation indicated raw oysters grown in South Australia as the cause of both ST417 and ST50 V. parahaemolyticus infections across Australia. Previous V. parahaemolyticus outbreaks have reported single-strain infections, predominantly by using traditional O and K serotyping methods (8,25). Longitudinal studies in Asia have identified a range of strains within a region (28), and other reports have highlighted highly virulent pandemic strains (e.g., ST36) detected across a widening international geographic range (1,13). Although minimal data are available in Australia regarding V. parahaemolyticus strains linked to locally acquired cases, recent increased use of genomic methods and practices will likely change that. The emergence of WGS characterization in Australia will also contribute to global knowledge regarding emerging and pathogenic strains. The presence of the trh virulence gene in both strains within this outbreak—and the additional tdh gene in the ST50 case isolates—correlates with prior literature noting that the presence of those virulence genes contributes to clinical symptoms but that both genes are not required to cause illness (29).
International risk assessments have been conducted for V. parahaemolyticus in seafood, noting the pathogenicity of the organism, the growth of V. parahaemolyticus increasing with increased water temperatures, and the need for strict postharvest controls to reduce the risk for foodborne disease (6,12). Outbreaks have occurred more frequently during warmer months (3) and at times when seawater temperatures have increased (8,20) or other environmental factors have had an influence (e.g., El Niño events or decreases in salinity) (10,30). In response to the outbreak we have described, oyster growers implemented postharvest controls through a Vibrio control program, where oysters were placed under active refrigeration. General trends of increased sea surface temperatures in Australia (31) and the seasonal occurrence of the Leeuwin current, which brings warm tropical waters to Western and South Australia (31), potentially created favorable conditions for growth of V. parahaemolyticus in South Australia oyster-growing bays. We believe that further research would improve understanding of risk factors for V. parahaemolyticus outbreaks in Australia’s prone regions, including ongoing environmental surveillance at harvest sites to monitor seawater temperatures, salinity, and harvest conditions, as well as at points along the storage and transport chain to consumers.
We noted that many outbreak case-patients in this study consumed medication that reduces stomach acid, a finding noted in previous studies that investigated risk for V. parahaemolyticus and other bacterial gastroenteric infections (32,33). Gastric acidity also decreases with age (34); therefore, infection susceptibility could increase with age, which is consistent with our observed median case patient age of 52 years. The fact that a large portion of our case-patients were older adults might also be related to food consumption patterns in the general population of Australia, where mollusks are less commonly eaten by children compared with adults (35). The outbreak we studied showed higher severity of illness than some previous outbreaks; for example, we noted 13% of case patients hospitalized and a single case with septicemia, compared with a study that investigated an outbreak associated with Alaska oysters, where there were no hospitalizations (8). Conversely, we noted a lower hospitalization rate for case-patients (13%) compared with a longer-term study that reported a hospitalization rate of 44% (9). Individual factors and the pathogenicity of different strains could affect disease severity in outbreaks.
The first limitation of our investigation is that culture for V. parahaemolyticus is not always attempted on diarrheal samples in diagnostic laboratories in Australia, and V. parahaemolyticus targets are often omitted in routine fecal multiplex PCR kits employed for direct detection of enteropathogens. Also, there was likely underreporting of cases because V. parahaemolyticus is not a notifiable condition in all jurisdictions in Australia. However, public health laboratories were contacted by their respective health departments and asked to provide information on enteric V. parahaemolyticus cases. Further typing of strains by WGS is also not consistently conducted across Australia and sometimes must be specifically requested if an outbreak is suspected. During this outbreak, some requests for further typing were made several weeks after the initial isolation, at which point no specimens were available for shipment to the public health laboratories. Because of incomplete typing of all case isolates in this outbreak, some cases might have been of a different ST and might not have been specifically linked to the current outbreak. In addition, V. parahaemolyticus outbreak cases we studied coincided with a national surge in SARS-CoV-2 infections in Australia, putting strain on public health resources. Therefore, not all case-patients were able to be interviewed, and not all isolates were able to be further typed.
There were also limitations in the traceback of oysters within a complex supply chain, including distributors and retailers receiving stock from multiple growers and different growing regions, leading to potential mixing of stock, some incomplete records and invoicing, and case-patients having multiple exposures to oysters within their incubation period. Mixing of oyster stock could also have contributed to the identification of multiple strains of V. parahaemolyticus in cases included in this outbreak. Although traceback was unable to be completed for all cases, oysters consumed by most casepatients were traced to at least the harvest area.
In conclusion, evidence for the source of this outbreak was strong, considering the oyster consumption among case-patients, traceback of the source of oysters consumed by case-patients, and identification of the same strain of V. parahaemolyticus in both oysters and case patients. The reduction in cases of V. parahaemolyticus after the recall of oysters and wide implementation of Vibrio control programs supports this evidence. This outbreak of V. parahaemolyticus associated with consumption of Australia-grown oysters, largely consumed raw, has led to improvements in postproduction control and traceability in the oyster industry in South Australia. The outbreak also spotlighted the virulent potential of V. parahaemolyticus and the value in distinguishing it as a nationally notifiable disease in Australia. Improved surveillance data, including strain identification from a wider range of regions, and a clearer understanding of underreporting have been highlighted by the World Health Organization as priorities for improving risk assessment processes for V. parahaemolyticus (22). Increased surveillance across all jurisdictions in Australia would improve outbreak detection and ensure a prompt and coordinated public health response.
Dr Fearnley is an infectious disease epidemiologist, working in the OzFoodNet network of epidemiologists in Australia. Her research interests include foodborne and zoonotic diseases and enteric disease modeling.
Acknowledgments
The authors thank the extended outbreak investigation team for contributions to this investigation, including the public health officers, food regulators, primary industry regulators, diagnostic laboratories, and public health laboratories in all jurisdictions. We thank specifically SA Pathology, PathWest Laboratory Medicine WA, Institute of Clinical Pathology and Medical Research-NSW Health Pathology, and Public Health Microbiology Forensic and Scientific Services (Queensland), Microbiological Diagnostic Unit. We also thank the AusTrakka National Analysis team as well as the OzFoodNet Network, which is funded by the Australian Government Department of Health and Aged Care. We extend thanks also to Food Standards Australia and New Zealand and the Department of Agriculture, Fisheries, and Forestry.
Ethics approval was not required for this study because the outbreak investigation was conducted under the relevant state, territory, and national public health legislation for investigation and response to acute public health events.
References
- Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, et al. Vibrio spp. infections. Nat Rev Dis Primers. 2018;4:8. DOIPubMedGoogle Scholar
- Heymann DL. Control of communicable diseases manual, 20th edition. Washington: American Public Health Association; 2015.
- Daniels NA, MacKinnon L, Bishop R, Altekruse S, Ray B, Hammond RM, et al. Vibrio parahaemolyticus infections in the United States, 1973-1998. J Infect Dis. 2000;181:1661–6. DOIPubMedGoogle Scholar
- Mannas H, Mimouni R, Chaouqy N, Hamadi F, Martinez-Urtaza J. Occurrence of Vibrio and Salmonella species in mussels (Mytilus galloprovincialis) collected along the Moroccan Atlantic coast. Springerplus. 2014;3:265. DOIPubMedGoogle Scholar
- Lopez-Joven C, de Blas I, Furones MD, Roque A. Prevalences of pathogenic and non-pathogenic Vibrio parahaemolyticus in mollusks from the Spanish Mediterranean Coast. Front Microbiol. 2015;6:736. DOIPubMedGoogle Scholar
- Wang D, Flint SH, Palmer JS, Gagic D, Fletcher GC, On SLW. Global expansion of Vibrio parahaemolyticus threatens the seafood industry: perspective on controlling its biofilm formation. Lebensm Wiss Technol. 2022;158:
113182 . DOIGoogle Scholar - González-Escalona N, Cachicas V, Acevedo C, Rioseco ML, Vergara JA, Cabello F, et al. Vibrio parahaemolyticus diarrhea, Chile, 1998 and 2004. Emerg Infect Dis. 2005;11:129–31. DOIPubMedGoogle Scholar
- McLaughlin JB, DePaola A, Bopp CA, Martinek KA, Napolilli NP, Allison CG, et al. Outbreak of Vibrio parahaemolyticus gastroenteritis associated with Alaskan oysters. N Engl J Med. 2005;353:1463–70. DOIPubMedGoogle Scholar
- Wu Y, Wen J, Ma Y, Ma X, Chen Y. Epidemiology of foodborne disease outbreaks caused by Vibrio parahaemolyticus, China, 2003–2008. Food Control. 2014;46:197–202. DOIGoogle Scholar
- Daniels NA, Ray B, Easton A, Marano N, Kahn E, McShan AL II, et al. Emergence of a new Vibrio parahaemolyticus serotype in raw oysters: A prevention quandary. JAMA. 2000;284:1541–5. DOIPubMedGoogle Scholar
- Newton A, Kendall M, Vugia DJ, Henao OL, Mahon BE. Increasing rates of vibriosis in the United States, 1996-2010: review of surveillance data from 2 systems. Clin Infect Dis. 2012;54(Suppl 5):S391–5. DOIPubMedGoogle Scholar
- Food and Agriculture Organization of the United Nations/World Health Organization. Risk assessment of Vibrio parahaemolyticus in seafood: Interpretative summary and technical report. Microbiological Risk Assessment Series No. 16. Rome: The Organizations; 2011 [cited 2022 Jul 28]. https://www.who.int/publications/i/item/9789241548175
- Martinez-Urtaza J, van Aerle R, Abanto M, Haendiges J, Myers RA, Trinanes J, et al. Genomic variation and evolution of Vibrio parahaemolyticus ST36 over the course of a transcontinental epidemic expansion. MBio. 2017;8:e01425–17. DOIPubMedGoogle Scholar
- Food and Drug Administration. U.S. Department of Health and Human Services, 2005. Quantitative risk assessment on the public health impact of pathogenic Vibrio parahaemolyticus in raw oysters. [cited 2022 Jun 7]. https://www.fda.gov/food/cfsan-risk-safety-assessments/quantitative-risk-assessment-public-health-impact-pathogenic-vibrio-parahaemolyticus-raw-oysters
- Harlock M, Quinn S, Turnbull AR. Emergence of non-choleragenic Vibrio infections in Australia. Commun Dis Intell (2018). 2022;46:46. DOIPubMedGoogle Scholar
- Australian Government Department of Health and Aged Care. 2023. OzFoodNet network. [cited 2023 Oct 6]. https://www.health.gov.au/our-work/ozfoodnet-network
- Jolley KA, Maiden MCJ. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11:595. DOIPubMedGoogle Scholar
- Hoang T, da Silva AG, Jennison AV, Williamson DA, Howden BP, Seemann T. AusTrakka: Fast-tracking nationalized genomics surveillance in response to the COVID-19 pandemic. Nat Commun. 2022;13:865. DOIPubMedGoogle Scholar
- Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3. DOIPubMedGoogle Scholar
- Taylor M, Cheng J, Sharma D, Bitzikos O, Gustafson R, Fyfe M, et al. Outbreak of Vibrio parahaemolyticus associated with consumption of raw oysters in Canada, 2015. Foodborne Pathog Dis. 2018;15:554–9. DOIPubMedGoogle Scholar
- Martinez-Urtaza J, Powell A, Jansa J, Rey JLC, Montero OP, Campello MG, et al. Epidemiological investigation of a foodborne outbreak in Spain associated with U.S. West Coast genotypes of Vibrio parahaemolyticus. Springerplus. 2016;5:87. DOIPubMedGoogle Scholar
- Food and Agriculture Organization of the United Nations/World Health Organization. Advances in science and risk assessment tools for Vibrio parahaemolyticus and V. vulnificus associated with seafood. Meeting report. Microbiological Risk Assessment Series No. 35. Rome: The Organizations; 2021 [cited 2022 Jun 7]
- Lopatek M, Wieczorek K, Osek J. Prevalence and antimicrobial resistance of Vibrio parahaemolyticus isolated from raw shellfish in Poland. J Food Prot. 2015;78:1029–33. DOIPubMedGoogle Scholar
- Cruz CD, Hedderley D, Fletcher GC. Long-term study of Vibrio parahaemolyticus prevalence and distribution in New Zealand shellfish. Appl Environ Microbiol. 2015;81:2320–7. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention (CDC). Vibrio parahaemolyticus infections associated with consumption of raw shellfish—three states, 2006. MMWR Morb Mortal Wkly Rep. 2006;55:854–6.PubMedGoogle Scholar
- Garbuglia AR, Bruni R, Villano U, Vairo F, Lapa D, Madonna E, et al.; The Other Members Of The Hev Outbreak Working Group. Hepatitis E outbreak in the central part of Italy sustained by multiple HEV genotype 3 strains, June–December 2019. Viruses. 2021;13:1159. DOIPubMedGoogle Scholar
- European Centre for Disease Prevention and Control. Multi-country outbreak of multiple Salmonella enterica serotypes linked to imported sesame-based products—14 October 2021. European Food Safety Authority, 2021 [cited 2024 June 19] https://www.ecdc.europa.eu/sites/default/files/documents/ROA_S%20Mbandaka_S%20Havana_UI-716_14-October-2021.pdf
- Li Y, Xie X, Shi X, Lin Y, Qiu Y, Mou J, et al. Vibrio parahaemolyticus, southern coastal region of China, 2007–2012. Emerg Infect Dis. 2014;20:685–8. DOIPubMedGoogle Scholar
- Raghunath P. Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front Microbiol. 2015;5:805. DOIPubMedGoogle Scholar
- Gonzalez-Escalona N, Gavilan RG, Toro M, Zamudio ML, Martinez-Urtaza J. Outbreak of Vibrio parahaemolyticus sequence type 120, Peru, 2009. Emerg Infect Dis. 2016;22:1235–7. DOIPubMedGoogle Scholar
- Bureau of Meteorology. State of the climate report, 2022 [cited 2022 Jun 7] http://www.bom.gov.au/state-of-the-climate/oceans.shtml
- Hassing RJ, Verbon A, de Visser H, Hofman A, Stricker BH. Proton pump inhibitors and gastroenteritis. Eur J Epidemiol. 2016;31:1057–63. DOIPubMedGoogle Scholar
- Cribb DM, Varrone L, Wallace RL, McLure AT, Smith JJ, Stafford RJ, et al. Risk factors for campylobacteriosis in Australia: outcomes of a 2018-2019 case-control study. BMC Infect Dis. 2022;22:586. DOIPubMedGoogle Scholar
- Koo J, Marshall DL, DePaola A. Antacid increases survival of Vibrio vulnificus and Vibrio vulnificus phage in a gastrointestinal model. Appl Environ Microbiol. 2001;67:2895–902. DOIPubMedGoogle Scholar
- Australian Bureau of Statistics. Australian Health Survey: Nutrition First results—foods and nutrients, 2011–2012 [cited 2024 Jan 18]. https://www.abs.gov.au/statistics/health/health-conditions-and-risks/australian-health-survey-nutrition-first-results-foods-and-nutrients/latest-release#data-downloads
Figures
Tables
Cite This ArticleOriginal Publication Date: October 16, 2024
Table of Contents – Volume 30, Number 11—November 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Emily Fearnley, SA Health, PO Box 14 Rundle Mall, Adelaide, South Australia 5000, Australia
Top