Volume 30, Number 11—November 2024
Research
Antiviral Susceptibility of Swine-Origin Influenza A Viruses Isolated from Humans, United States
Figure
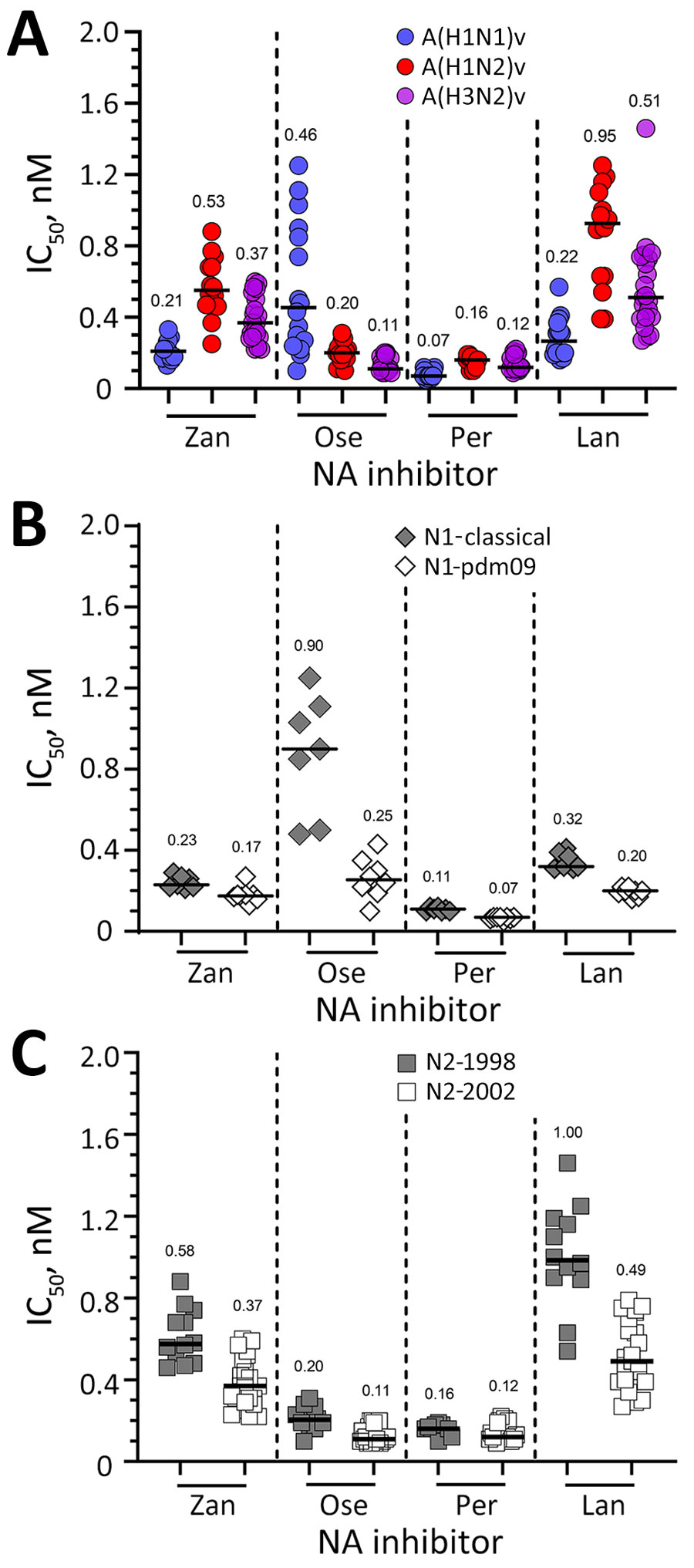
Figure. Susceptibility of variant viruses to NA inhibitors based on subtype and NA lineage in study of antiviral susceptibility of swine-origin influenza A viruses isolated from humans, United States. A) Susceptibility of A(H1N1)v (n = 15), A(H1N2)v (n = 14), and A(H3N2)v (n = 21) viruses to NA inhibitors determined in a fluorescence-based assay (29). The IC50s of viruses lacking known or suspected molecular markers that reduce inhibition by NA inhibitors were used to calculate the subtype-specific median IC50s (baseline susceptibility). B, C) Effect of NA lineage on inhibition by NA inhibitors. IC50s obtained in NA inhibition assay were grouped according to their respective NA lineage: N1-classical (n = 7, closed diamond), N1-pdm09 (n = 8, open diamond), N2-1998 (n = 12, closed square), or N2-2002 (n = 23, open square). Horizontal bars and numbers indicate median IC50s. IC50, 50% inhibitory concentration; Lan, laninamivir; NA, neuraminidase; Ose, oseltamivir; Per, peramivir; Zan, zanamivir.
References
- Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, et al. Review of influenza A virus in swine worldwide: a call for increased surveillance and research. Zoonoses Public Health. 2014;61:4–17. DOIPubMedGoogle Scholar
- Scholtissek C, Bürger H, Kistner O, Shortridge KF. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology. 1985;147:287–94. DOIPubMedGoogle Scholar
- Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–73. DOIPubMedGoogle Scholar
- Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, et al. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–6. DOIPubMedGoogle Scholar
- Olsen CW. The emergence of novel swine influenza viruses in North America. Virus Res. 2002;85:199–210. DOIPubMedGoogle Scholar
- World Health Organization. International health regulations, 2nd edition. Geneva: The Organization; 2005.
- Council of State and Territorial Epidemiologists. Council of State and Territorial Epidemiologists Position statement: national reporting for initial detections of novel influenza A viruses [cited 2024 Jun 17]. https://cdn.ymaws.com/www.cste.org/resource/resmgr/PS/07-ID-01.pdf
- Shu B, Garten R, Emery S, Balish A, Cooper L, Sessions W, et al. Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990-2010. Virology. 2012;422:151–60. DOIPubMedGoogle Scholar
- Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, et al.; Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. DOIPubMedGoogle Scholar
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science. 2009;325:197–201. DOIPubMedGoogle Scholar
- Anderson TK, Chang J, Arendsee ZW, Venkatesh D, Souza CK, Kimble JB, et al. Swine influenza A viruses and the tangled relationship with humans. Cold Spring Harb Perspect Med. 2021;11:
a038737 . DOIPubMedGoogle Scholar - Markin A, Ciacci Zanella G, Arendsee ZW, Zhang J, Krueger KM, Gauger PC, et al. Reverse-zoonoses of 2009 H1N1 pandemic influenza A viruses and evolution in United States swine results in viruses with zoonotic potential. PLoS Pathog. 2023;19:
e1011476 . DOIPubMedGoogle Scholar - World Health Organization. Standardization of terminology for the influenza virus variants infecting humans: update [cited 2024 Jun 17]. https://www.who.int/publications/m/item/standardization-of-terminology-for-the-influenza-virus-variants-infecting-humans-update
- Jhung MA, Epperson S, Biggerstaff M, Allen D, Balish A, Barnes N, et al. Outbreak of variant influenza A(H3N2) virus in the United States. Clin Infect Dis. 2013;57:1703–12. DOIPubMedGoogle Scholar
- Sleeman K, Mishin VP, Guo Z, Garten RJ, Balish A, Fry AM, et al. Antiviral susceptibility of variant influenza A(H3N2)v viruses isolated in the United States from 2011 to 2013. Antimicrob Agents Chemother. 2014;58:2045–51. DOIPubMedGoogle Scholar
- Nelson MI, Stratton J, Killian ML, Janas-Martindale A, Vincent AL. Continual reintroduction of human pandemic H1N1 influenza A viruses into swine in the United States, 2009 to 2014. J Virol. 2015;89:6218–26. DOIPubMedGoogle Scholar
- Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM, Webster RG. Evolution of swine H3N2 influenza viruses in the United States. J Virol. 2000;74:8243–51. DOIPubMedGoogle Scholar
- Rajão DS, Walia RR, Campbell B, Gauger PC, Janas-Martindale A, Killian ML, et al. Reassortment between swine H3N2 and 2009 pandemic H1N1 in the United States resulted in influenza A viruses with diverse genetic constellations with variable virulence in pigs. J Virol. 2017;91:e01763–16. DOIPubMedGoogle Scholar
- Sharma A, Zeller MA, Li G, Harmon KM, Zhang J, Hoang H, et al. Detection of live attenuated influenza vaccine virus and evidence of reassortment in the U.S. swine population. J Vet Diagn Invest. 2020;32:301–11. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Novel influenza A virus infections [cited 2024 Apr 24]. https://gis.cdc.gov/grasp/fluview/Novel_Influenza.html
- Cox NJ, Trock SC, Burke SA. Pandemic preparedness and the Influenza Risk Assessment Tool (IRAT). Curr Top Microbiol Immunol. 2014;385:119–36. DOIPubMedGoogle Scholar
- World Health Organization. Tool for influenza pandemic risk assessment (TIPRA), version 2 release. January 2020 [cited 2024 Jun 17]. https://www.who.int/publications/i/item/tool-for-influenza-pandemic-risk-assessment-(tipra)-2nd-edition
- Jones JC, Yen HL, Adams P, Armstrong K, Govorkova EA. Influenza antivirals and their role in pandemic preparedness. Antiviral Res. 2023;210:
105499 . DOIPubMedGoogle Scholar - Hurt AC. The epidemiology and spread of drug resistant human influenza viruses. Curr Opin Virol. 2014;8:22–9. DOIPubMedGoogle Scholar
- Takashita E, Ejima M, Itoh R, Miura M, Ohnishi A, Nishimura H, et al. A community cluster of influenza A(H1N1)pdm09 virus exhibiting cross-resistance to oseltamivir and peramivir in Japan, November to December 2013. Euro Surveill. 2014;19:20666. DOIPubMedGoogle Scholar
- World Health Organization. Global Influenza Surveillance Network: manual for the laboratory diagnosis and virological surveillance of influenza. Geneva: The Organization; 2011.
- Shepard SS, Meno S, Bahl J, Wilson MM, Barnes J, Neuhaus E. Viral deep sequencing needs an adaptive approach: IRMA, the iterative refinement meta-assembler. BMC Genomics. 2016;17:708. DOIPubMedGoogle Scholar
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. DOIPubMedGoogle Scholar
- Okomo-Adhiambo M, Sleeman K, Ballenger K, Nguyen HT, Mishin VP, Sheu TG, et al. Neuraminidase inhibitor susceptibility testing in human influenza viruses: a laboratory surveillance perspective. Viruses. 2010;2:2269–89. DOIPubMedGoogle Scholar
- Patel MC, Flanigan D, Feng C, Chesnokov A, Nguyen HT, Elal AA, et al. An optimized cell-based assay to assess influenza virus replication by measuring neuraminidase activity and its applications for virological surveillance. Antiviral Res. 2022;208:
105457 . DOIPubMedGoogle Scholar - World Health Organization. Meetings of the WHO working group on surveillance of influenza antiviral susceptibility – Geneva, November 2011 and June 2012. Wkly Epidemiol Rec. 2012;87:369–74.PubMedGoogle Scholar
- World Health Organization. Laboratory methodologies for testing the antiviral susceptibility of influenza viruses: neuraminidase inhibitor (NAI) [cited 2024 Jun 17]. https://www.who.int/teams/global-influenza-programme/laboratory-network/quality-assurance/antiviral-susceptibility-influenza/neuraminidase-inhibitor
- World Health Organization. Laboratory methodologies for testing the antiviral susceptibility of influenza viruses: polymerase acidic (PA) inhibitor, baloxavir [cited 2024 Jun 17]. https://www.who.int/teams/global-influenza-programme/laboratory-network/quality-assurance/antiviral-susceptibility-influenza/polymerase-acidic-protein-inhibitor
- Boltz DA, Douangngeun B, Phommachanh P, Sinthasak S, Mondry R, Obert C, et al. Emergence of H5N1 avian influenza viruses with reduced sensitivity to neuraminidase inhibitors and novel reassortants in Lao People’s Democratic Republic. J Gen Virol. 2010;91:949–59. DOIPubMedGoogle Scholar
- Hurt AC, Lee RT, Leang SK, Cui L, Deng YM, Phuah SP, et al. Increased detection in Australia and Singapore of a novel influenza A(H1N1)2009 variant with reduced oseltamivir and zanamivir sensitivity due to a S247N neuraminidase mutation. Euro Surveill. 2011;16:19884. DOIPubMedGoogle Scholar
- Mishin VP, Patel MC, Chesnokov A, De La Cruz J, Nguyen HT, Lollis L, et al. Susceptibility of influenza A, B, C, and D viruses to baloxavir. Emerg Infect Dis. 2019;25:1969–72. DOIPubMedGoogle Scholar
- Omoto S, Speranzini V, Hashimoto T, Noshi T, Yamaguchi H, Kawai M, et al. Characterization of influenza virus variants induced by treatment with the endonuclease inhibitor baloxavir marboxil. Sci Rep. 2018;8:9633. DOIPubMedGoogle Scholar
- Stevaert A, Dallocchio R, Dessì A, Pala N, Rogolino D, Sechi M, et al. Mutational analysis of the binding pockets of the diketo acid inhibitor L-742,001 in the influenza virus PA endonuclease. J Virol. 2013;87:10524–38. DOIPubMedGoogle Scholar
- Govorkova EA, Takashita E, Daniels RS, Fujisaki S, Presser LD, Patel MC, et al. Global update on the susceptibilities of human influenza viruses to neuraminidase inhibitors and the cap-dependent endonuclease inhibitor baloxavir, 2018-2020. Antiviral Res. 2022;200:
105281 . DOIPubMedGoogle Scholar - Gubareva LV, Mishin VP, Patel MC, Chesnokov A, Nguyen HT, De La Cruz J, et al. Assessing baloxavir susceptibility of influenza viruses circulating in the United States during the 2016/17 and 2017/18 seasons. Euro Surveill. 2019;24:
1800666 . DOIPubMedGoogle Scholar - Takashita E, Daniels RS, Fujisaki S, Gregory V, Gubareva LV, Huang W, et al. Global update on the susceptibilities of human influenza viruses to neuraminidase inhibitors and the cap-dependent endonuclease inhibitor baloxavir, 2017-2018. Antiviral Res. 2020;175:
104718 . DOIPubMedGoogle Scholar - Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–6. DOIPubMedGoogle Scholar
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337:1343–8. DOIPubMedGoogle Scholar
- Leung RC, Ip JD, Chen LL, Chan WM, To KK. Global emergence of neuraminidase inhibitor-resistant influenza A(H1N1)pdm09 viruses with I223V and S247N mutations: implications for antiviral resistance monitoring. Lancet Microbe. 2024;5:627–8. DOIPubMedGoogle Scholar
- Patel M, Nguyen HT, Pascua PNQ, Gao R, Steel J, Kondor RJ, et al. Multicountry spread of influenza A(H1N1)pdm09 viruses with reduced inhibition by oseltamivir, May 2023–February 2024. Emerg Infect Dis. 2024;30:1410–5. DOIPubMedGoogle Scholar
- Takashita E, Fujisaki S, Morita H, Nagata S, Miura H, Matsuura Y, et al. A community cluster of influenza A(H3N2) virus infection with reduced susceptibility to baloxavir due to a PA E199G substitution in Japan, February to March 2023. Euro Surveill. 2023;28:
2300501 . DOIPubMedGoogle Scholar - World Health Organization. 2024. Influenza at the human-animal interface summary and risk assessment, 26 February 2024 [cited 2024 Jun 17]. https://www.who.int/publications/m/item/influenza-at-the-human-animal-interface-summary-and-assessment-26--february-2024
- Cogdale J, Kele B, Myers R, Harvey R, Lofts A, Mikaiel T, et al.; Influenza A(H1N2)v Incident Management Team. A case of swine influenza A(H1N2)v in England, November 2023. Euro Surveill. 2024;29:
2400002 . DOIPubMedGoogle Scholar
1These authors contributed equally to this article.