Volume 30, Number 12—December 2024
Research
Human Circovirus in Patients with Hepatitis, Hong Kong
Figure 4
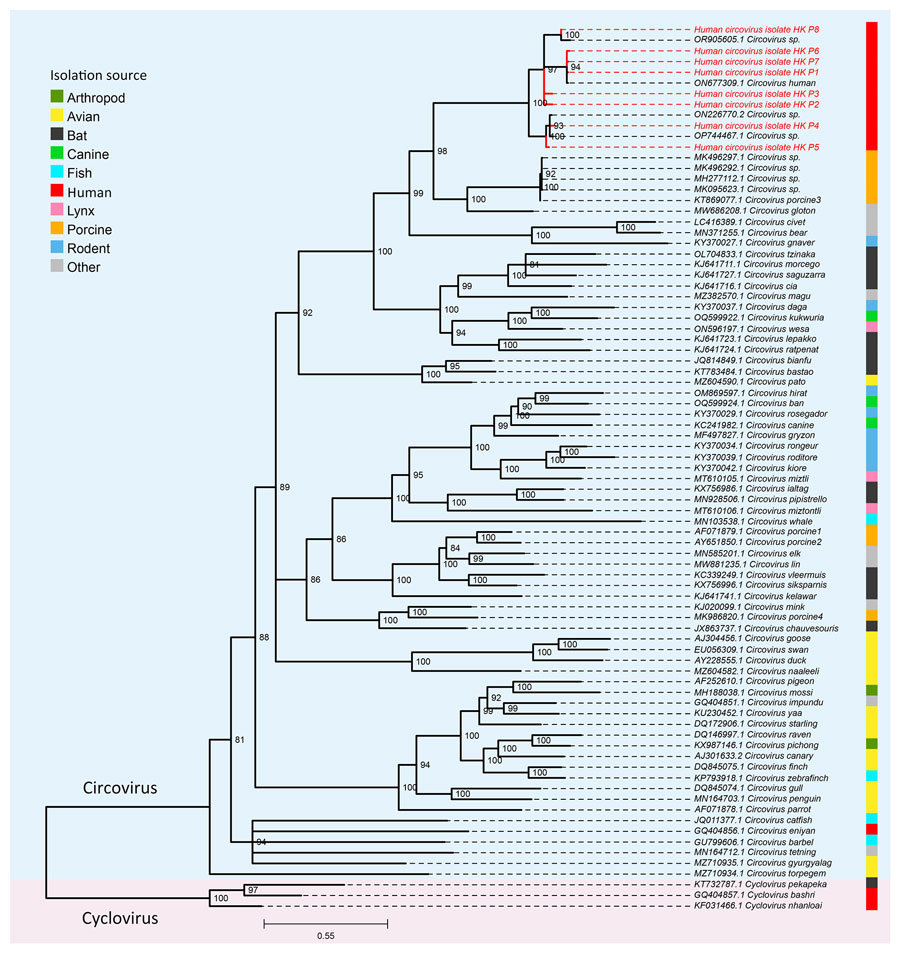
Figure 4. Phylogenetic tree of representative circoviruses and human circoviruses from study of human circovirus in patients with hepatitis, Hong Kong. Maximum-likelihood phylogenetic tree was inferred from a multiple sequence alignment of complete representative Circovirus genomes and a mixture of full-length and partial genome sequences from this study (red text) using IQ-TREE (16). Three representative species of Cyclovirus were used as an outgroup for rooting the tree. Branch supports were assessed using the Shimodaira-Hasegawa–like approximate likelihood ratio test with 10,000 bootstrap replicates. Branches with <80% support were collapsed. Scale bar indicates nucleotide substitutions per site.
References
- Bernal W, Auzinger G, Dhawan A, Wendon J. Acute liver failure. Lancet. 2010;376:190–201. DOIPubMedGoogle Scholar
- Ho A, Orton R, Tayler R, Asamaphan P, Herder V, Davis C, et al.; DIAMONDS Consortium; ISARIC4C Investigators. Adeno-associated virus 2 infection in children with non-A-E hepatitis. Nature. 2023;617:555–63. DOIPubMedGoogle Scholar
- Sridhar S, Yip CCY, Wu S, Cai J, Zhang AJ, Leung KH, et al. Rat hepatitis E virus as cause of persistent hepatitis after liver transplant. Emerg Infect Dis. 2018;24:2241–50. DOIPubMedGoogle Scholar
- Rivero-Juarez A, Frias M, Perez AB, Pineda JA, Reina G, Fuentes-Lopez A, et al.; HEPAVIR and GEHEP-014 Study Groups. Orthohepevirus C infection as an emerging cause of acute hepatitis in Spain: First report in Europe. J Hepatol. 2022;77:326–31. DOIPubMedGoogle Scholar
- Sridhar S, Yip CCY, Lo KHY, Wu S, Situ J, Chew NFS, et al. Hepatitis E virus species C infection in humans, Hong Kong. Clin Infect Dis. 2022;75:288–96. DOIPubMedGoogle Scholar
- Li Y, Cao L, Ye M, Xu R, Chen X, Ma Y, et al. Plasma virome reveals blooms and transmission of anellovirus in intravenous drug users with HIV-1, HCV, and/or HBV infections. Microbiol Spectr. 2022;10:
e0144722 . DOIPubMedGoogle Scholar - Pérot P, Fourgeaud J, Rouzaud C, Regnault B, Da Rocha N, Fontaine H, et al. Circovirus hepatitis infection in heart-lung transplant patient, France. Emerg Infect Dis. 2023;29:286–93. DOIPubMedGoogle Scholar
- Varsani A, Harrach B, Roumagnac P, Benkő M, Breitbart M, Delwart E, et al. 2024 taxonomy update for the family Circoviridae. Arch Virol. 2024;169:176. DOIPubMedGoogle Scholar
- Li Y, Zhang P, Ye M, Tian RR, Li N, Cao L, et al. Novel circovirus in blood from intravenous drug users, Yunnan, China. Emerg Infect Dis. 2023;29:1015–9. DOIPubMedGoogle Scholar
- Wong SSY, Yip CCY, Sridhar S, Leung KH, Cheng AKW, Fung AMY, et al. Comparative evaluation of a laboratory-developed real-time PCR assay and RealStar® Adenovirus PCR Kit for quantitative detection of human adenovirus. Virol J. 2018;15:149. DOIPubMedGoogle Scholar
- Yip CCY, Sridhar S, Cheng AKW, Fung AMY, Cheng VCC, Chan KH, et al. Comparative evaluation of a laboratory developed real-time PCR assay and the RealStar® HHV-6 PCR Kit for quantitative detection of human herpesvirus 6. J Virol Methods. 2017;246:112–6. DOIPubMedGoogle Scholar
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. DOIPubMedGoogle Scholar
- Katoh K, Frith MC. Adding unaligned sequences into an existing alignment using MAFFT and LAST. Bioinformatics. 2012;28:3144–6. DOIPubMedGoogle Scholar
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. DOIPubMedGoogle Scholar
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. DOIPubMedGoogle Scholar
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. DOIPubMedGoogle Scholar
- Bianchini G, Sánchez-Baracaldo P. TreeViewer: Flexible, modular software to visualise and manipulate phylogenetic trees. Ecol Evol. 2024;14:
e10873 . DOIPubMedGoogle Scholar - Sridhar S, Situ J, Cai JP, Yip CC, Wu S, Zhang AJ, et al. Multimodal investigation of rat hepatitis E virus antigenicity: Implications for infection, diagnostics, and vaccine efficacy. J Hepatol. 2021;74:1315–24. DOIPubMedGoogle Scholar
- Sridhar S, Yip CC, Wu S, Chew NF, Leung KH, Chan JF, et al. Transmission of rat hepatitis E virus infection to humans in Hong Kong: a clinical and epidemiological analysis. Hepatology. 2021;73:10–22. DOIPubMedGoogle Scholar
- Yip CCY, Sridhar S, Leung KH, Cheng AKW, Chan KH, Chan JFW, et al. Evaluation of RealStar Alpha Herpesvirus PCR Kit for detection of HSV-1, HSV-2, and VZV in clinical specimens. BioMed Res Int. 2019;2019:
5715180 . DOIPubMedGoogle Scholar - Sridhar S, Chung TWH, Chan JFW, Cheng VCC, Lau SKP, Yuen KY, et al. Emergence of cytomegalovirus mononucleosis syndrome among young adults in Hong Kong linked to falling seroprevalence: results of a 14-year seroepidemiological study. Open Forum Infect Dis. 2018;5:
ofy262 . DOIPubMedGoogle Scholar - Smith C, Tsang J, Beagley L, Chua D, Lee V, Li V, et al. Effective treatment of metastatic forms of Epstein-Barr virus-associated nasopharyngeal carcinoma with a novel adenovirus-based adoptive immunotherapy. Cancer Res. 2012;72:1116–25. DOIPubMedGoogle Scholar
- Maggi F, Pifferi M, Fornai C, Andreoli E, Tempestini E, Vatteroni M, et al. TT virus in the nasal secretions of children with acute respiratory diseases: relations to viremia and disease severity. J Virol. 2003;77:2418–25. DOIPubMedGoogle Scholar
- Kwo PY, Cohen SM, Lim JK. ACG clinical guideline: evaluation of abnormal liver chemistries. Am J Gastroenterol. 2017;112:18–35. DOIPubMedGoogle Scholar
- Pérot P, Da Rocha N, Farcet MR, Kreil TR, Eloit M. Human Circovirus is not detected in plasma pools for fractionation. Transfusion. 2024;64:16–8. DOIPubMedGoogle Scholar
- Sridhar S, To KK, Chan JF, Lau SK, Woo PC, Yuen KY. A systematic approach to novel virus discovery in emerging infectious disease outbreaks. J Mol Diagn. 2015;17:230–41. DOIPubMedGoogle Scholar
- Smits SL, Zijlstra EE, van Hellemond JJ, Schapendonk CM, Bodewes R, Schürch AC, et al. Novel cyclovirus in human cerebrospinal fluid, Malawi, 2010-2011. Emerg Infect Dis. 2013;19:1511–3. DOIPubMedGoogle Scholar
- Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, et al. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology. 2013;436:295–303. DOIPubMedGoogle Scholar
- Deinhardt F, Holmes AW, Capps RB, Popper H. Studies on the transmission of human viral hepatitis to marmoset monkeys. I. Transmission of disease, serial passages, and description of liver lesions. J Exp Med. 1967;125:673–88. DOIPubMedGoogle Scholar
- Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92–7. DOIPubMedGoogle Scholar
- Sridhar S, Yip CCY, Chew NFS, Wu S, Leung KH, Chan JFW, et al. Epidemiological and clinical characteristics of human hepegivirus 1 infection in patients with hepatitis C. Open Forum Infect Dis. 2019;6:
ofz329 . DOIPubMedGoogle Scholar - Berg MG, Lee D, Coller K, Frankel M, Aronsohn A, Cheng K, et al. Discovery of a novel human pegivirus in blood associated with hepatitis C virus co-infection. PLoS Pathog. 2015;11:
e1005325 . DOIPubMedGoogle Scholar - Li IW, To KK, Tang BS, Chan KH, Hui CK, Cheng VC, et al. Human metapneumovirus infection in an immunocompetent adult presenting as mononucleosis-like illness. J Infect. 2008;56:389–92. DOIPubMedGoogle Scholar
- Chiu KH, Wong SC, Tam AR, Sridhar S, Yip CC, Chan KH, et al. The first case of monkeypox in Hong Kong presenting as infectious mononucleosis-like syndrome. Emerg Microbes Infect. 2023;12:
2146910 . DOIPubMedGoogle Scholar - Muñoz-Gómez S, Cunha BA. Parvovirus B19 mimicking Epstein-Barr virus infectious mononucleosis in an adult. Am J Med. 2013;126:e7–8. DOIPubMedGoogle Scholar
- Hamelin B, Pérot P, Pichler I, Haslbauer JD, Hardy D, Hing D, et al. Circovirus hepatitis in immunocompromised patient, Switzerland. Emerg Infect Dis. 2024;30:2140–4. DOIPubMedGoogle Scholar
- da Silva RR, da Silva DF, da Silva VH, de Castro AMMG. Porcine circovirus 3: a new challenge to explore. Front Vet Sci. 2024;10:
1266499 . DOIPubMedGoogle Scholar - Chen D, Zhang L, Xu S. Pathogenicity and immune modulation of porcine circovirus 3. Front Vet Sci. 2023;10:
1280177 . DOIPubMedGoogle Scholar - Krüger L, Längin M, Reichart B, Fiebig U, Kristiansen Y, Prinz C, et al. Transmission of porcine circovirus 3 (PCV3) by xenotransplantation of pig hearts into baboons. Viruses. 2019;11:650. DOIPubMedGoogle Scholar
1These authors contributed equally to this article.