Volume 30, Number 2—February 2024
Dispatch
Experimental SARS-CoV-2 Infection of Elk and Mule Deer
Cite This Article
Citation for Media
Abstract
To assess the susceptibility of elk (Cervus canadensis) and mule deer (Odocoileus hemionus) to SARS-CoV-2, we performed experimental infections in both species. Elk did not shed infectious virus but mounted low-level serologic responses. Mule deer shed and transmitted virus and mounted pronounced serologic responses and thus could play a role in SARS-CoV-2 epidemiology.
Natural infections and experimental studies have indicated that diverse species of mammals can be infected with SARS-CoV-2 (1). The angiotensin-converting enzyme 2 receptor of white-tailed deer is closely homologous to that of humans. Angiotensin-converting enzyme 2 modeling studies predict that cervids, including sika deer (Cervus nippon), reindeer (Rangifer tarandus), and Père David’s deer (Elaphurus davidianus), are susceptible to SARS-CoV-2 (2–4). White-tailed deer are susceptible to experimental infection with SARS-CoV-2, subsequently shedding infectious virus and infecting naive conspecifics (5–7). Surveillance studies have demonstrated SARS-CoV-2 infection in free-ranging and captive white-tailed deer in the United States and Canada, determined by viral RNA detection, antibodies to SARS-CoV-2, or virus isolation (8–11). After their displacement in humans, the Alpha and Delta variants of concern persisted in white-tailed deer populations (12,13). Because SARS-CoV-2 is likely to have repeatedly spilled over from humans to white-tailed deer and circulated within North America deer populations, we assessed susceptibility of elk (Cervus canadensis) and mule deer (Odocoileus hemionus) to the Delta variant of SARS-CoV-2. All animal work was approved by the Colorado State University (Fort Collins, Colorado, USA) Institutional Animal Care and Use Committee.
We studied 6 weanling elk (all female) and 6 yearling mule deer (5 female, 1 male). Animals were procured from private vendors and group housed (3 individuals of the same species/room) in an animal Biosafety Level 3 facility at Colorado State University. Before study commencement, all animals were seronegative for SARS-CoV-2.
We passaged the Delta variant of SARS-CoV-2 (BEI Resources, National Institute of Allergy and Infectious Diseases, National Institutes of Health: isolate hCoV-19/USA/MD-HP05647/2021 [lineage B.1.617.2]) 1 time in Vero cells. We then sequenced the isolate by using a next-generation pipeline and detected a single synonymous consensus change at amino acid 410 in nonstructural protein 14 (C to T transition) (GenBank accession no. OR758451) (14). We then intranasally inoculated 2 animals/room with 3.7–4.5 log10 PFU of virus diluted in phosphate-buffered saline (confirmed by back-titration of inoculum on Vero cells); the third animal in each room served as a contact.
We assessed the animals daily for signs of clinical disease (e.g., lethargy, anorexia, nasal discharge, sneezing, coughing, and dyspnea). One mule deer (no. 3) was tachypneic and coughing at arrival, and clinical signs continued throughout the week-long acclimation period until euthanasia at 3 days postinoculation (dpi) according to the study schedule. Because that respiratory pattern was present before study commencement, we did not attribute it to SARS-CoV-2 infection. No other clinical signs were observed in any animal.
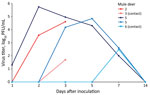
At 0, 1, 2, 3, 5, 7, and 14 dpi, we collected oral, nasal, and rectal swab samples. Because no elk shed infectious virus orally or nasally, we did not assess elk rectal swab samples. We performed reverse transcription PCR on elk oral and nasal swab samples collected through 7 dpi. We recovered SARS-CoV-2 RNA from samples from 3 directly inoculated elk collected at 1–5 dpi; cycle threshold values for all 3 were >28 (Table 1). Plaque assay showed that 3 of the 4 directly inoculated mule deer shed infectious virus orally and nasally (Figures 1, 2). Oral shedding of virus commenced at either 2 or 3 dpi and resolved by 7 dpi for directly inoculated mule deer. One contact mule deer shed virus orally at 7 dpi (Figure 1). Nasal shedding of virus was more staggered; inoculated animals initially shed virus at 1, 2, or 3 dpi, continuing through 7 dpi (in the 2 direct inoculants remaining at that time). For contact mule deer, only 1 nasal sample per animal was positive, collected at either 3 or 7 dpi (Figure 2). Infectious virus was not recovered from any mule deer rectal samples.
At 3 dpi, we euthanized and necropsied animals from 1 room of each species (n = 3; 2 inoculants and 1 contact) and collected tissues (nasal turbinates, trachea, heart, lung, liver, spleen, kidney, small intestine) for virus isolation and histopathologic examination. Although we did not detect infectious virus in any elk tissues, we recovered infectious virus from the nasal turbinates and trachea of 1 directly inoculated mule deer.
The remaining animals were maintained until 21 dpi, when they were euthanized and underwent necropsy with the same tissues collected into formalin. Blood from those animals was collected weekly and evaluated for a serologic response to SARS-CoV-2 by plaque reduction neutralization test (15). A low-level antibody response developed in both directly inoculated elk; peak neutralizing titers were 1:20 at 21 dpi. The contact elk did not seroconvert. Virus-neutralizing antibodies developed in all mule deer held until 21 dpi; peak titers reached >1:1,280 (Table 2).
At necropsy, we observed no gross lesions in any animals. A veterinary pathologist evaluated all respiratory tissues from all mule deer. The tracheas of all mule deer were histologically unremarkable, but multifocal accumulations of mononuclear leukocytes in the absence of frank inflammation were noted in the nasal turbinates, lungs, or both from 5 mule deer. Elk tissues did not undergo histologic evaluation.
If wildlife populations serve as maintenance hosts for SARS-CoV-2, the implications could be substantial. The persistence of virus variants already displaced in the human population, virus evolution, and spillback into a human have all been suggested to have occurred in white-tailed deer populations (11,12), although it is still unclear whether those deer will serve as maintenance hosts of the virus. Evaluating the susceptibility of other cervid species to SARS-CoV-2 will help direct surveillance efforts among free-ranging wildlife, which are key to understanding the epidemiology of SARS-CoV-2 and implementing control measures.
We used the Delta variant of SARS-CoV-2 to challenge animals in this experiment on the basis of evidence that this variant of concern was prevalent in white-tailed deer populations (13). Our results indicate that although elk seem to be minimally susceptible to infection with the Delta variant, mule deer are highly susceptible and capable of transmitting the virus. Inoculated elk showed no clinical signs, did not shed infectious virus, and mounted low-level humoral titers. The genomic RNA recovered from elk could represent residual inoculum. Infection in mule deer was subclinical, and although immune activation in the absence of frank inflammation was observed in respiratory tissues from 5 of the 6 animals, that finding may or may not be linked to SARS-CoV-2 infection. Of note, all mule deer used in this study were incidentally tested to assess their chronic wasting disease (CWD) status. Animals no. 1 and 3 were CWD positive, although it is unlikely that a concurrent infection with CWD greatly affected their susceptibility to infection with SARS-CoV-2 because only 1 of those animals became infected with SARS-CoV-2 while the other was the sole mule deer in our study that did not.
Experimental infection of elk and mule deer with SARS-CoV-2 revealed that although elk are minimally susceptible to infection, mule deer become infected, shed infectious virus, and can infect naive contacts. Mule deer are less widely distributed than white-tailed deer but still represent a population of cervids that is frequently in contact with humans and domestic animals. Therefore, susceptibility of mule deer provides yet another potential source for SARS-CoV-2 spillover or spillback. At this time, there is no evidence that wildlife are a significant source of SARS-CoV-2 exposure for humans, but the potential for this virus to become established in novel host species could lead to virus evolution in which novel variants may arise. Therefore, continued surveillance of species at risk, such as white-tailed and mule deer, is needed to detect any variants quickly and prevent transmission.
Dr. Porter is a science fellow with the National Wildlife Research Center at the US Department of Agriculture. Her research interests include the pathogenesis and transmission of infectious diseases.
Acknowledgments
We thank and recognize the US Department of Agriculture, Animal and Plant Health Inspection Service Science Fellows Program for supporting the salary of S.M.P. We are very grateful to Tracy Nichols, Colorado and Kansas state veterinarians, and Colorado Parks and Wildlife for logistical support. Special thanks to Jeremy Ellis for technical assistance; Susan Shriner for consulting; McKinzee Barker, Elizabeth Lawrence, and Hannah Sueper for their help with animal handling; and Richard Bowen for facilities support.
This work was supported by internal funding from Colorado State University and the US Department of Agriculture, Animal and Plant Health Inspection Service. This study was directly funded by the American Rescue Plan Act provision to conduct monitoring and surveillance of animals susceptible to SARS-CoV-2.
References
- Frazzini S, Amadori M, Turin L, Riva F. SARS CoV-2 infections in animals, two years into the pandemic. Arch Virol. 2022;167:2503–17. DOIPubMedGoogle Scholar
- Lopes LR, Carvalho de Lucca Pina N, da Silva AC Junior, Bandiera-Paiva P. Evolutionary analysis of mammalian ACE2 and the key residues involved in binding to the spike protein revealed potential SARS-CoV-2 hosts. Journal of Medical Microbiology and Infectious Diseases. 2022;10:1–9. DOIGoogle Scholar
- Fischhoff IR, Castellanos AA, Rodrigues JPGLM, Varsani A, Han BA. Predicting the zoonotic capacity of mammals to transmit SARS-CoV-2. Proc Biol Sci. 2021;288:
20211651 . DOIPubMedGoogle Scholar - Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci U S A. 2020;117:22311–22. DOIPubMedGoogle Scholar
- Palmer MV, Martins M, Falkenberg S, Buckley A, Caserta LC, Mitchell PK, et al. Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. J Virol. 2021;95:e00083–21. DOIPubMedGoogle Scholar
- Martins M, Boggiatto PM, Buckley A, Cassmann ED, Falkenberg S, Caserta LC, et al. From Deer-to-Deer: SARS-CoV-2 is efficiently transmitted and presents broad tissue tropism and replication sites in white-tailed deer. PLoS Pathog. 2022;18:
e1010197 . DOIPubMedGoogle Scholar - Cool K, Gaudreault NN, Morozov I, Trujillo JD, Meekins DA, McDowell C, et al. Infection and transmission of ancestral SARS-CoV-2 and its alpha variant in pregnant white-tailed deer. Emerg Microbes Infect. 2022;11:95–112. DOIPubMedGoogle Scholar
- Chandler JC, Bevins SN, Ellis JW, Linder TJ, Tell RM, Jenkins-Moore M, et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc Natl Acad Sci U S A. 2021;118:
e2114828118 . DOIPubMedGoogle Scholar - Hale VL, Dennis PM, McBride DS, Nolting JM, Madden C, Huey D, et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature. 2022;602:481–6. DOIPubMedGoogle Scholar
- Kuchipudi SV, Surendran-Nair M, Ruden RM, Yon M, Nissly RH, Vandegrift KJ, et al. Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer. Proc Natl Acad Sci U S A. 2022;119:
e2121644119 . DOIPubMedGoogle Scholar - Pickering B, Lung O, Maguire F, Kruczkiewicz P, Kotwa JD, Buchanan T, et al. Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission. Nat Microbiol. 2022;7:2011–24. DOIPubMedGoogle Scholar
- Marques AD, Sherrill-Mix S, Everett JK, Adhikari H, Reddy S, Ellis JC, et al. Multiple introductions of SARS-CoV-2 Alpha and Delta variants into white-tailed deer in Pennsylvania. MBio. 2022;13:
e0210122 . DOIPubMedGoogle Scholar - McBride DS, Garushyants SK, Franks J, Magee AF, Overend SH, Huey D, et al. Accelerated evolution of SARS-CoV-2 in free-ranging white-tailed deer. Nat Commun. 2023;14:5105. DOIPubMedGoogle Scholar
- Marano JM, Weger-Lucarelli J. Replication in the presence of dengue convalescent serum impacts Zika virus neutralization sensitivity and fitness. Front Cell Infect Microbiol. 2023;13:
1130749 . DOIPubMedGoogle Scholar - Bosco-Lauth AM, Hartwig AE, Porter SM, Gordy PW, Nehring M, Byas AD, et al. Experimental infection of domestic dogs and cats with SARS-CoV-2: Pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci U S A. 2020;117:26382–8. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: January 12, 2024
1These senior authors contributed equally to this article.
Table of Contents – Volume 30, Number 2—February 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Angela Bosco-Lauth, Department of Biomedical Sciences, Colorado State University, 1683 Campus Delivery, Fort Collins, CO 80523, USA
Top