Volume 30, Number 4—April 2024
Research
Clostridium butyricum Bacteremia Associated with Probiotic Use, Japan
Figure
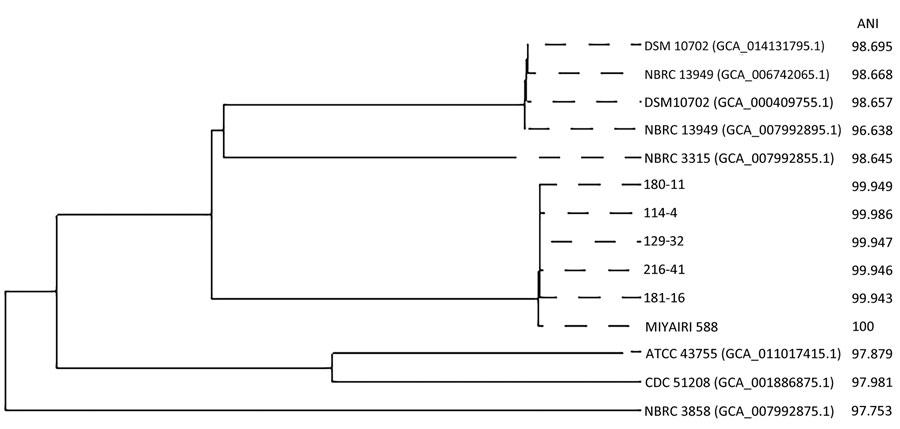
Figure. Phylogenetic tree reflecting the relationship between Clostridium butyricum MIYAIRI 588, clinical isolates of C. butyricum, and 8 reference strains based on data from a single-institute, retrospective study, Osaka University Hospital, Japan. Note: 114-4, 129-32, 180-11, 181-16, and 216-41 represent strain numbers of clinical isolates of C. butyricum. MIYAIRI 588 indicates C. butyricum MIYAIRI 588. DSM10702 (GCA_014131795.1), NBRC 13949 (GCA_006742065.1), DSM 10702 (GCA_000409755.1), NBRC 13949 (GCA_007992895.1), NBRC 3315 (GCA_007992895.1), ATCC 43755 (GCA_011017415.1), CDC 51208 (GCA_001886875.1), and NBRC 3858 (GCA_007992875.1) represent 8 reference strains. ANI was calculated using FastANI (31). ANI, average nucleotide identity.
References
- Kerry RG, Patra JK, Gouda S, Park Y, Shin HS, Das G. Benefaction of probiotics for human health: A review. Yao Wu Shi Pin Fen Xi. 2018;26:927–39. DOIPubMedGoogle Scholar
- Collinson S, Deans A, Padua-Zamora A, Gregorio GV, Li C, Dans LF, et al. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst Rev. 2020;12:
CD003048 .PubMedGoogle Scholar - Zhang T, Zhang C, Zhang J, Sun F, Duan L. Efficacy of probiotics for irritable bowel syndrome: a systematic review and network meta-analysis. Front Cell Infect Microbiol. 2022;12:
859967 . DOIPubMedGoogle Scholar - Hempel S, Newberry SJ, Maher AR, Wang Z, Miles JN, Shanman R, et al. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. JAMA. 2012;307:1959–69. DOIPubMedGoogle Scholar
- Zhu J, Pitre T, Ching C, Zeraatkar D, Gruchy S. Safety and efficacy of probiotic supplements as adjunctive therapies in patients with COVID-19: A systematic review and meta-analysis. PLoS One. 2023;18:
e0278356 . DOIPubMedGoogle Scholar - Kerry RG, Patra JK, Gouda S, Park Y, Shin HS, Das G. Benefaction of probiotics for human health: A review. Yao Wu Shi Pin Fen Xi. 2018;26:927–39. DOIPubMedGoogle Scholar
- Mountzouris KC, McCartney AL, Gibson GR. Intestinal microflora of human infants and current trends for its nutritional modulation. Br J Nutr. 2002;87:405–20. DOIPubMedGoogle Scholar
- Tran NT, Li Z, Ma H, Zhang Y, Zheng H, Gong Y, et al. Clostridium butyricum: a promising probiotic confers positive health benefits in aquatic animals. Rev Aquacult. 2020;12:2573–89. DOIGoogle Scholar
- Finegold SM, Sutter VL, Mathisen GE. Normal indigenous intestinal flora. In: Hentges DJ, editor. Human Intestinal Microflora in Health and Disease. New York: Elsevier Inc. 1983. p. 1:3–31.
- Ghoddusi HB, Sherburn R. Preliminary study on the isolation of Clostridium butyricum strains from natural sources in the UK and screening the isolates for presence of the type E botulinal toxin gene. Int J Food Microbiol. 2010;142:202–6. DOIPubMedGoogle Scholar
- Isa K, Oka K, Beauchamp N, Sato M, Wada K, Ohtani K, et al. Safety assessment of the Clostridium butyricum MIYAIRI 588® probiotic strain including evaluation of antimicrobial sensitivity and presence of Clostridium toxin genes in vitro and teratogenicity in vivo. Hum Exp Toxicol. 2016;35:818–32. DOIPubMedGoogle Scholar
- Maeda A, Ishii K, Tanaka M, Mikami Y, Arai T. KM1, a bacteriophage of Clostridium butyricum. Microbiology. 1986;132:2271–5. DOIGoogle Scholar
- Oka K, McCartney E, Ariyoshi T, Kudo H, Vilá B, de Jong L, et al. In vivo safety evaluation of the Clostridium butyricum MIYAIRI 588 strain in broilers, piglets, and turkeys. Toxicol Res Appl. 2019;3. DOIGoogle Scholar
- Hagihara M, Ariyoshi T, Kuroki Y, Eguchi S, Higashi S, Mori T, et al. Clostridium butyricum enhances colonization resistance against Clostridioides difficile by metabolic and immune modulation. Sci Rep. 2021;11:15007. DOIPubMedGoogle Scholar
- Takahashi M, Taguchi H, Yamaguchi H, Osaki T, Sakazaki R, Kamiya S. [Antagonistic interaction between Clostridium butyricum and enterohemorrhagic Escherichia coli O157:H7] [in Japanese]. Kansenshogaku Zasshi. 1999;73:7–14. DOIPubMedGoogle Scholar
- Seki H, Shiohara M, Matsumura T, Miyagawa N, Tanaka M, Komiyama A, et al. Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr Int. 2003;45:86–90. DOIPubMedGoogle Scholar
- Muldrew KL. Rapidly fatal postlaparoscopic liver infection from the rarely isolated species Clostridium butyricum. Case Rep Infect Dis. 2020;2020:
1839456 . DOIPubMedGoogle Scholar - Smith MF, Borriello SP, Clayden GS, Casewell MW. Clinical and bacteriological findings in necrotising enterocolitis: a controlled study. J Infect. 1980;2:23–31. DOIPubMedGoogle Scholar
- Sato Y, Kujirai D, Emoto K, Yagami T, Yamada T, Izumi M, et al. Necrotizing enterocolitis associated with Clostridium butyricum in a Japanese man. Acute Med Surg. 2018;5:194–8. DOIPubMedGoogle Scholar
- Cassir N, Benamar S, La Scola B. Clostridium butyricum: from beneficial to a new emerging pathogen. Clin Microbiol Infect. 2016;22:37–45. DOIPubMedGoogle Scholar
- Cassir N, Benamar S, Khalil JB, Croce O, Saint-Faust M, Jacquot A, et al. Clostridium butyricum strains and dysbiosis linked to necrotizing enterocolitis in preterm neonates. Clin Infect Dis. 2015;61:1107–15. DOIPubMedGoogle Scholar
- Shimura M, Mizuma M, Nakagawa K, Aoki S, Miura T, Takadate T, et al. Probiotic-related bacteremia after major hepatectomy for biliary cancer: a report of two cases. Surg Case Rep. 2021;7:133. DOIPubMedGoogle Scholar
- Ishikawa K, Hasegawa R, Shibutani K, Mikami Y, Kawai F, Matsuo T, et al. Probiotic-related Clostridium butyricum bacteremia: a case report and literature review. Anaerobe. 2023;83:
102770 . DOIPubMedGoogle Scholar - von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP; STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007;147:573–7. DOIPubMedGoogle Scholar
- Sulaiman IM, Miranda N, Simpson S. MALDI-TOF mass spectrometry and 16S rRNA gene sequence analysis for the identification of foodborne Clostridium spp. J AOAC Int. 2021;104:1381–8. DOIPubMedGoogle Scholar
- Quan H, Li B, Couris CM, Fushimi K, Graham P, Hider P, et al. Updating and validating the Charlson comorbidity index and score for risk adjustment in hospital discharge abstracts using data from 6 countries. Am J Epidemiol. 2011;173:676–82. DOIPubMedGoogle Scholar
- Wilkins T, Sequoia J. Probiotics for gastrointestinal conditions: a summary of the evidence. Am Fam Physician. 2017;96:170–8.PubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 33rd ed (M100-ED33). Wayne (PA): The Institute; 2023.
- Mory F, Lozniewski A, Bland S, Sedallian A, Grollier G, Girard-Pipau F, et al. Survey of anaerobic susceptibility patterns: a French multicentre study. Int J Antimicrob Agents. 1998;10:229–36. DOIPubMedGoogle Scholar
- Hecht DW. Anaerobes: antibiotic resistance, clinical significance, and the role of susceptibility testing. Anaerobe. 2006;12:115–21. DOIPubMedGoogle Scholar
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. DOIPubMedGoogle Scholar
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. DOIPubMedGoogle Scholar
- Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. DOIPubMedGoogle Scholar
- Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–9. DOIPubMedGoogle Scholar
- Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. DOIPubMedGoogle Scholar
- Costa RL, Moreira J, Lorenzo A, Lamas CC. Infectious complications following probiotic ingestion: a potentially underestimated problem? A systematic review of reports and case series. BMC Complement Altern Med. 2018;18:329. DOIPubMedGoogle Scholar
- Linz B, Windsor HM, McGraw JJ, Hansen LM, Gajewski JP, Tomsho LP, et al. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat Commun. 2014;5:4165. DOIPubMedGoogle Scholar
- Hassoun-Kheir N, Snitser O, Hussein K, Rabino G, Eluk O, Warman S, et al. Concordance between epidemiological evaluation of probability of transmission and whole genome sequence relatedness among hospitalized patients acquiring Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae. Clin Microbiol Infect. 2021;27:468.e1–7. DOIPubMedGoogle Scholar
- Goldenberg JZ, Lytvyn L, Steurich J, Parkin P, Mahant S, Johnston BC. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev. 2015;12:
CD004827 . DOIPubMedGoogle Scholar - Goldenberg JZ, Ma SS, Saxton JD, Martzen MR, Vandvik PO, Thorlund K, et al. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev. 2013;5:
CD006095 . DOIGoogle Scholar - Xu J, Ma R, Chen LF, Zhao LJ, Chen K, Zhang RB. Effects of probiotic therapy on hepatic encephalopathy in patients with liver cirrhosis: an updated meta-analysis of six randomized controlled trials. Hepatobiliary Pancreat Dis Int. 2014;13:354–60. DOIPubMedGoogle Scholar
- Naidoo K, Gordon M, Fagbemi AO, Thomas AG, Akobeng AK. Probiotics for maintenance of remission in ulcerative colitis. Cochrane Database Syst Rev. 2011;12:
CD007443 . DOIPubMedGoogle Scholar
Page created: February 06, 2024
Page updated: March 20, 2024
Page reviewed: March 20, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.