Volume 30, Number 4—April 2024
Research
Clostridium butyricum Bacteremia Associated with Probiotic Use, Japan
Abstract
Clostridium butyricum, a probiotic commonly prescribed in Asia, most notably as MIYA-BM (Miyarisan Pharmaceutical Co., Ltd.; https://www.miyarisan.com), occasionally leads to bacteremia. The prevalence and characteristics of C. butyricum bacteremia and its bacteriologic and genetic underpinnings remain unknown. We retrospectively investigated patients admitted to Osaka University Hospital during September 2011–February 2023. Whole-genome sequencing revealed 5 (0.08%) cases of C. butyricum bacteremia among 6,576 case-patients who had blood cultures positive for any bacteria. Four patients consumed MIYA-BM, and 1 patient consumed a different C. butyricum-containing probiotic. Most patients had compromised immune systems, and common symptoms included fever and abdominal distress. One patient died of nonocclusive mesenteric ischemia. Sequencing results confirmed that all identified C. butyricum bacteremia strains were probiotic derivatives. Our findings underscore the risk for bacteremia resulting from probiotic use, especially in hospitalized patients, necessitating judicious prescription practices.
Probiotics have emerged as agents that improve a wide range of conditions and provide essential ingredients for potential health benefits. Probiotics exhibit a diverse array of effects by engaging in competitive interactions with pathogenic microbial communities, competing for binding sites, helping exclude pathogens, and triggering activation of specific genes within and beyond the host’s intestinal tract. This process, in turn, stimulates, regulates, and controls the immune response (1). Probiotics have been found to be effective not only in managing conditions such as acute gastroenteritis (2) and irritable bowel syndrome (3) but also in preventing antibiotic-associated diarrhea (4) and even in alleviating symptoms associated with COVID-19 (5).
Clostridium butyricum is a strictly anaerobic, gram-positive, spore-forming bacillus named for its capacity to produce high amounts of butyric acid. C. butyricum was first isolated from the intestines of pigs by Prazmowski in 1880 (6), and several strains of C. butyricum have been reported from various environments in humans (7) and animals (8). C. butyricum has been detected in the gut of ≈20% of human adults (9). Moreover, C. butyricum strains were detected in >30% of environmental samples tested (10). Some strains of C. butyricum are currently used as probiotics and have beneficial effects on humans and animals. One strain of C. butyricum, known as C. butyricum MIYAIRI 588 (CBM 588), can be found in pharmaceutical probiotics, such as MIYA-BM (Miyarisan Pharmaceutical Co., Ltd., https://www.miyarisan.com), one of the most commonly prescribed probiotics in Japan. CBM 588 has been described as a unique, nongenetically modified strain that does not naturally produce toxins (11) or cause disease owing to its susceptibility to the KM1 bacteriophage (12). Several confirmatory factors underpin this characterization: it exhibits no propensity for antibiotic resistance transfer, it is devoid of plasmids bearing mobile genetic elements, and it does not possess genes or produce substances related to clostridial toxins, including botulinum neurotoxins A, B, E, and F, or the Clostridium perfringens toxins α, β, and ε. Genomic scrutiny of CBM 588 revealed no indicators of pathogenic traits or hemolytic capabilities (13). Numerous studies have substantiated the effectiveness of CBM 588, and various animal model experiments have demonstrated its capacity to inhibit the colonization of Clostridioides difficile (14) and prevent enterohemorrhagic Escherichia coli O157 infection (15). Human studies have confirmed that CBM 588 prevents antibiotic-associated diarrhea (16). In the medical context in Japan, CBM 588 has been prescribed not only for its expected effectiveness as a conventional probiotic but also for the prophylaxis of the diseases we have listed.
There are, however, other strains of C. butyricum that are involved in infectious diseases (17–21). A few case reports have noted the development of C. butyricum bacteremia in patients taking probiotics, although strain definition tests using whole-genome sequencing were not conducted (22,23). Bacteremia caused by C. butyricum is a rare condition, and the prevalence, clinical features, and bacteriologic and genetic origins of the strains are unknown. We conducted a single-center, retrospective study of cases of bacteremia caused by C. butyricum in Japan to shed light on this clinical event.
Study Design
We conducted a retrospective cohort study at Osaka University Hospital, a 1,086-bed facility in Osaka, Japan. Our study followed the Strengthening the Reporting of Observational Studies in Epidemiology statement for reporting observational studies (24). The Institutional Review Board of Osaka University Hospital approved the study protocol (number 22584(T1)).
Patients and Baseline Characteristics
To identify cases of C. butyricum bacteremia, we reviewed all cases of positive blood culture results for any bacteria that occurred during September 19, 2011–February 5, 2023, from the Laboratory for Clinical Investigation database at Osaka University Hospital. We defined C. butyricum bacteremia as cases in which C. butyricum was detected in >1 sets of blood cultures. We used MALDI Biotyper (Bruker, https://www.bruker.com/en) to identify C. butyricum (25). The data we extracted from medical records encompassed such parameters as age; sex; conditions necessitating hospitalization; underlying diseases; placement of a central venous catheter or a peripherally inserted central catheter; presence of polymicrobial bacteremia, including identification of microorganisms other than C. butyricum; symptoms at onset; and the updated Charlson Comorbidity Index at the time of bacteremia diagnosis, which was evaluated for every patient (26). In addition, for patients who were prescribed MIYA-BM, we checked the MIYA-BM consumption at the point of diagnosis and confirmed the duration of MIYA-BM prescription. We also identified whether MIYA-BM was used for specific reasons in these patients. We defined specific reasons for use of MIYA-BM as treatment for diarrhea, concurrent antibiotic use, or medical history of C. difficile infection (CDI), ulcerative colitis, hepatic encephalopathy, or a combination of those conditions. Our investigation involved a detailed evaluation of electronic medical records, which included symptoms of diarrhea occurring ≥3 times/day, concurrent antibiotic use, and medical history of CDI, ulcerative colitis, or hepatic encephalopathy. Finally, we extracted data on the etiology of bacteremia, antibiotic treatment regimens, and mortality within 90 days.
Microbiologic Information
We determined the MICs for penicillin, ampicillin, cefotaxime, ceftriaxone, cefmetazole, imipenem, meropenem, sulbactam/ampicillin, clavulanic acid/amoxicillin, tazobactam/piperacillin, clindamycin, moxifloxacin, and metronidazole for C. butyricum by using the agar dilution method on Brucella agar medium supplemented with 0.5% sheep’s blood. Assays to gauge susceptibility followed the guidelines set by the Clinical Laboratory Standards Institute, tailored for anaerobes (28). We assessed the homogeneity of antibiotic susceptibility between the clinical strains and 3 medicinal strains from different lot numbers to evaluate the comparability of their antibiotic susceptibility.
Whole-Genome Sequencing Analysis
We conducted whole-genome analysis of all strains of C. butyricum obtained from blood cultures. In addition, we analyzed C. butyricum extracted from MIYA-BM tablets. We then investigated the genetic homology between those strains by evaluating the number of single-nucleotide polymorphisms (SNPs) or insertion/deletion genetic variants between clinical strains and the strain from the MIYA-BM tablets. Finally, we conducted a genomic comparison between clinical isolates of C. butyricum, the CBM 588 strain, and other strains of the same species. For the comparison, in addition to the reference strain CDC 51208, we selected 7 strains with fully sequenced genomes that are stored in a bioresource repository.
We detected 5 blood culture–positive C. butyricum bacteremia cases (0.08%) (Table 1) from a total of 6,576 persons who had blood cultures positive for any bacteria (7,484 total clinical strains, including bacteria other than C. butyricum). Bacteremia developed in all 5 patients during hospitalization; 3 patients were women and 2 were men. Four patients were immunocompromised: 2 had undergone transplantation, 1 was undergoing chemotherapy for esophageal and gastric cancers, and 1 was receiving multiple immunosuppressive treatments for dermatomyositis. Two of the 5 patients also had end-stage kidney disease and were on dialysis. The Charlson Comorbidity Index scores ranged from 1 to 6 points for each patient. Three patients underwent catheterization with either a central venous catheter or a peripherally inserted central catheter. Four patients were taking prescribed MIYA-BM at the time of bacteremia diagnosis, and 1 patient (no. 2) had been prescribed a different probiotic containing C. butyricum 1 month before the diagnosis of bacteremia. All 4 patients taking MIYA-BM were prescribed it >1 week prior to hospitalization, and MIYA-BM was discontinued following the diagnosis of bacteremia in all these patients. Despite a detailed review of the medical records, we were unable to identify the specific reason for prescribing probiotics in 2 patients. All 5 patients had fever and abdominal symptoms, such as diarrhea and pain. One patient (no. 3) with nonocclusive mesenteric ischemia died within 90 days.
A consistent pattern of antibiotic susceptibility was observed in all clinical strains (Table 2). Moreover, those results were consistent with those of previous reports on the antibiotic susceptibility of C. butyricum. C. butyricum has been reported to be susceptible to penicillin, ampicillin, cefmetazole, imipenem, meropenem, clavulanic acid/amoxicillin, tazobactam/piperacillin, clindamycin, moxifloxacin, and metronidazole but resistant to cefotaxime and ceftriaxone (11,29,30).
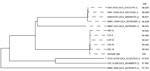
Whole-genome analysis of all 5 patient clinical strains revealed that they either exhibited complete homology or had a maximum divergence of only 19 mutations relative to CBM 588, which was extracted from the MIYA-BM tablets. This result indicates that all clinical strains had the same clone as the CBM 588 extracted from MIYA-BM (Table 3) (31–34). We performed genetic annotation of the detected mutations (Appendix Table). We performed phylogenetic analysis of C. butyricum by using the Type (Strain) Genome Server (35). All clinical isolates and probiotics strain were clustered on the same branches (Figure). Average nucleotide identity scores of clinical isolates against those of the probiotics strain were higher than against those of reference strains. This analysis further corroborated the genetic homology between all the clinical strains and the CBM 588 strain.
Our single-center, retrospective study determined that the prevalence of C. butyricum bacteremia was 0.08% among all cases with bacteria-positive whole-blood cultures and that all clinical strains were derived from the CBM 588 strain. Bacteremia developed in all patients during hospitalization. Out of 5 cases, 4 had received immunosuppressive treatment and 2 had intra-abdominal issues (1 case of esophageal and gastric cancer and 1 case of post–pancreas and kidney transplantation).
Ishikawa et al. reported a case series of 11 cases of C. butyricum bacteremia, including 3 self-experienced cases and 8 cases from a literature review (23). The study revealed that at least 8 cases developed bacteremia during their hospitalization for conditions unrelated to the bacteremia itself. Furthermore, most patients had intra-abdominal issues at the time of developing bacteremia. In 3 cases, C. butyricum bacteremia developed after intra-abdominal surgery. Among the 8 cases without intra-abdominal surgery, 6 cases occurred after various intra-abdominal conditions (2 cases of Crohn’s disease, 2 cases of gastrointestinal ulcers, 1 case of biliary tract infection, and 1 case of nonobstructive mesenteric ischemia). Our study results align with previous findings, emphasizing the need for vigilant monitoring of bacteremia development associated with probiotic use in patients with intra-abdominal issues or those undergoing immunosuppressive therapy during their hospitalization.
Our study revealed a high degree of genetic similarity between the strains of C. butyricum extracted from MIYA-BM tablets and clinical strains identified through genetic analysis, strongly supporting the definition of probiotic-related bacteremia in all our cases. Reports on probiotic-related bacteremia are scarce. Although systematic reviews of cases of bacteremia after probiotic use have been reported (36), to the best of our knowledge, no studies have evaluated the genetic similarities among these reports. Our study offers evidence supporting a direct causal relationship between probiotic prescription and bacteremia. Nonetheless, the patients we identified as nos. 1 and 2 present lingering challenges. We observed 19 differences in terms of SNPs between the strains found in the blood culture of patient 1 and the CBM 588 strain, which was relatively higher than that of the other patients. However, it is common to evaluate strain dissimilarity using fewer than 100 SNPs. Notably, rapidly growing bacteria, such as Helicobacter pylori, can accumulate ≈30 SNPs within 6 months of acute infection (37). In fact, some studies have established a genetic similarity cutoff of 80 for carbapenem-resistant Klebsiella pneumoniae (38), suggesting that the genetic dissimilarity observed in this case could be reasonably acceptable. We also considered the possibility that long-term oral administration of probiotics in the past could have led to genetic mutations in the CBM 588 strain within the bodies of patients we examined. Patient 2, who had been prescribed a different probiotic containing C. butyricum 1 month before the diagnosis of bacteremia, developed bacteremia caused by the CBM 588 strain. We considered 2 possibilities for this observation: the patient had previously taken MIYA-BM and it had colonized in the patient’s gastrointestinal tract, leading to an infection; or the C. butyricum present in the probiotics the patient was taking had genetic similarities to the CBM 588 strain.
Our findings also bring to light the potential adverse effects related to the inappropriate prescribing of probiotics. In all cases where MIYA-BM was prescribed, probiotics were administered for >1 week. However, after a comprehensive review of the detailed medical records, we were unable to identify the appropriate reasons for prescribing probiotics in half of the cases. Probiotics exhibit various therapeutic and preventive effects in different medical conditions, such as averting antibiotic-associated diarrhea (39) and CDI (40), preventing hepatic encephalopathy in patients with liver cirrhosis (41), and managing symptoms in patients with ulcerative colitis (42). Although probiotics may demonstrate effectiveness in such specialized clinical scenarios, those scenarios were not observed in the cases we studied, in which probiotics appeared to have been prescribed indiscriminately over an extended period.
One limitation of our study was that it was a single-center, retrospective investigation. Multicenter studies are needed to elucidate the prevalence of C. butyricum bacteremia and the genetic origin of the strains. Another limitation was that patient 1 showed improvement with ceftriaxone use, although C. butyricum is resistant to it. There is a possibility of contamination resulting from such factors as polymicrobial bacteremia and the absence of central venous catheterization. However, it cannot be ruled out that patients with concurrent sacral pressure ulcers are at risk of developing polymicrobial bacteremia, including C. butyricum bacteremia. Also, the duration of probiotic use for each case patient was based on information documented in their medical records, and the precise prescription durations were not always clear. However, the actual prescription periods must exceed the durations documented in the medical records, because the recorded periods represent at least the minimum assessable timeframe. Moreover, although specific reasons for probiotic prescription were not evident in the medical records, unique justifications may have existed. Nevertheless, it is crucial to note that none of the patients had a history of prior antibiotic use, CDI, irritable bowel syndrome, or liver cirrhosis. Hence, the need for prolonged administration exceeding 2 weeks for therapeutic purposes seems unlikely.
In conclusion, our study demonstrates that all clinical strains of C. butyricum identified in the positive blood cultures of the 5 cases we analyzed were derived from the strain found in probiotics. Although this type of bacteremia is rare, careful monitoring is essential when bacteremia is caused by probiotics. Clinicians must avoid long-term, inappropriate prescription of probiotics for hospitalized patients with multiple comorbidities, including immunosuppressive treatment and intraabdominal problems, to prevent bacteremia caused by probiotics.
Acknowledgments
This research was conducted as part of the All-Osaka U Research in “The Nippon Foundation– Osaka University Infectious Disease Response Project.”
Dr Sada is an endowed chair associate professor of the Department of Transformative Protection to Infectious Disease, Graduate School of Medicine, Osaka University, Osaka, Japan. His research interests include the epidemiology of bacteremia, infections caused by rare bacteria, and immunodeficiency-related infections.
References
- Kerry RG, Patra JK, Gouda S, Park Y, Shin HS, Das G. Benefaction of probiotics for human health: A review. Yao Wu Shi Pin Fen Xi. 2018;26:927–39. DOIPubMedGoogle Scholar
- Collinson S, Deans A, Padua-Zamora A, Gregorio GV, Li C, Dans LF, et al. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst Rev. 2020;12:
CD003048 .PubMedGoogle Scholar - Zhang T, Zhang C, Zhang J, Sun F, Duan L. Efficacy of probiotics for irritable bowel syndrome: a systematic review and network meta-analysis. Front Cell Infect Microbiol. 2022;12:
859967 . DOIPubMedGoogle Scholar - Hempel S, Newberry SJ, Maher AR, Wang Z, Miles JN, Shanman R, et al. Probiotics for the prevention and treatment of antibiotic-associated diarrhea: a systematic review and meta-analysis. JAMA. 2012;307:1959–69. DOIPubMedGoogle Scholar
- Zhu J, Pitre T, Ching C, Zeraatkar D, Gruchy S. Safety and efficacy of probiotic supplements as adjunctive therapies in patients with COVID-19: A systematic review and meta-analysis. PLoS One. 2023;18:
e0278356 . DOIPubMedGoogle Scholar - Kerry RG, Patra JK, Gouda S, Park Y, Shin HS, Das G. Benefaction of probiotics for human health: A review. Yao Wu Shi Pin Fen Xi. 2018;26:927–39. DOIPubMedGoogle Scholar
- Mountzouris KC, McCartney AL, Gibson GR. Intestinal microflora of human infants and current trends for its nutritional modulation. Br J Nutr. 2002;87:405–20. DOIPubMedGoogle Scholar
- Tran NT, Li Z, Ma H, Zhang Y, Zheng H, Gong Y, et al. Clostridium butyricum: a promising probiotic confers positive health benefits in aquatic animals. Rev Aquacult. 2020;12:2573–89. DOIGoogle Scholar
- Finegold SM, Sutter VL, Mathisen GE. Normal indigenous intestinal flora. In: Hentges DJ, editor. Human Intestinal Microflora in Health and Disease. New York: Elsevier Inc. 1983. p. 1:3–31.
- Ghoddusi HB, Sherburn R. Preliminary study on the isolation of Clostridium butyricum strains from natural sources in the UK and screening the isolates for presence of the type E botulinal toxin gene. Int J Food Microbiol. 2010;142:202–6. DOIPubMedGoogle Scholar
- Isa K, Oka K, Beauchamp N, Sato M, Wada K, Ohtani K, et al. Safety assessment of the Clostridium butyricum MIYAIRI 588® probiotic strain including evaluation of antimicrobial sensitivity and presence of Clostridium toxin genes in vitro and teratogenicity in vivo. Hum Exp Toxicol. 2016;35:818–32. DOIPubMedGoogle Scholar
- Maeda A, Ishii K, Tanaka M, Mikami Y, Arai T. KM1, a bacteriophage of Clostridium butyricum. Microbiology. 1986;132:2271–5. DOIGoogle Scholar
- Oka K, McCartney E, Ariyoshi T, Kudo H, Vilá B, de Jong L, et al. In vivo safety evaluation of the Clostridium butyricum MIYAIRI 588 strain in broilers, piglets, and turkeys. Toxicol Res Appl. 2019;3. DOIGoogle Scholar
- Hagihara M, Ariyoshi T, Kuroki Y, Eguchi S, Higashi S, Mori T, et al. Clostridium butyricum enhances colonization resistance against Clostridioides difficile by metabolic and immune modulation. Sci Rep. 2021;11:15007. DOIPubMedGoogle Scholar
- Takahashi M, Taguchi H, Yamaguchi H, Osaki T, Sakazaki R, Kamiya S. [Antagonistic interaction between Clostridium butyricum and enterohemorrhagic Escherichia coli O157:H7] [in Japanese]. Kansenshogaku Zasshi. 1999;73:7–14. DOIPubMedGoogle Scholar
- Seki H, Shiohara M, Matsumura T, Miyagawa N, Tanaka M, Komiyama A, et al. Prevention of antibiotic-associated diarrhea in children by Clostridium butyricum MIYAIRI. Pediatr Int. 2003;45:86–90. DOIPubMedGoogle Scholar
- Muldrew KL. Rapidly fatal postlaparoscopic liver infection from the rarely isolated species Clostridium butyricum. Case Rep Infect Dis. 2020;2020:
1839456 . DOIPubMedGoogle Scholar - Smith MF, Borriello SP, Clayden GS, Casewell MW. Clinical and bacteriological findings in necrotising enterocolitis: a controlled study. J Infect. 1980;2:23–31. DOIPubMedGoogle Scholar
- Sato Y, Kujirai D, Emoto K, Yagami T, Yamada T, Izumi M, et al. Necrotizing enterocolitis associated with Clostridium butyricum in a Japanese man. Acute Med Surg. 2018;5:194–8. DOIPubMedGoogle Scholar
- Cassir N, Benamar S, La Scola B. Clostridium butyricum: from beneficial to a new emerging pathogen. Clin Microbiol Infect. 2016;22:37–45. DOIPubMedGoogle Scholar
- Cassir N, Benamar S, Khalil JB, Croce O, Saint-Faust M, Jacquot A, et al. Clostridium butyricum strains and dysbiosis linked to necrotizing enterocolitis in preterm neonates. Clin Infect Dis. 2015;61:1107–15. DOIPubMedGoogle Scholar
- Shimura M, Mizuma M, Nakagawa K, Aoki S, Miura T, Takadate T, et al. Probiotic-related bacteremia after major hepatectomy for biliary cancer: a report of two cases. Surg Case Rep. 2021;7:133. DOIPubMedGoogle Scholar
- Ishikawa K, Hasegawa R, Shibutani K, Mikami Y, Kawai F, Matsuo T, et al. Probiotic-related Clostridium butyricum bacteremia: a case report and literature review. Anaerobe. 2023;83:
102770 . DOIPubMedGoogle Scholar - von Elm E, Altman DG, Egger M, Pocock SJ, Gøtzsche PC, Vandenbroucke JP; STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann Intern Med. 2007;147:573–7. DOIPubMedGoogle Scholar
- Sulaiman IM, Miranda N, Simpson S. MALDI-TOF mass spectrometry and 16S rRNA gene sequence analysis for the identification of foodborne Clostridium spp. J AOAC Int. 2021;104:1381–8. DOIPubMedGoogle Scholar
- Quan H, Li B, Couris CM, Fushimi K, Graham P, Hider P, et al. Updating and validating the Charlson comorbidity index and score for risk adjustment in hospital discharge abstracts using data from 6 countries. Am J Epidemiol. 2011;173:676–82. DOIPubMedGoogle Scholar
- Wilkins T, Sequoia J. Probiotics for gastrointestinal conditions: a summary of the evidence. Am Fam Physician. 2017;96:170–8.PubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing, 33rd ed (M100-ED33). Wayne (PA): The Institute; 2023.
- Mory F, Lozniewski A, Bland S, Sedallian A, Grollier G, Girard-Pipau F, et al. Survey of anaerobic susceptibility patterns: a French multicentre study. Int J Antimicrob Agents. 1998;10:229–36. DOIPubMedGoogle Scholar
- Hecht DW. Anaerobes: antibiotic resistance, clinical significance, and the role of susceptibility testing. Anaerobe. 2006;12:115–21. DOIPubMedGoogle Scholar
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. DOIPubMedGoogle Scholar
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. DOIPubMedGoogle Scholar
- Cingolani P, Platts A, Wang L, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012;6:80–92. DOIPubMedGoogle Scholar
- Tanizawa Y, Fujisawa T, Nakamura Y. DFAST: a flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–9. DOIPubMedGoogle Scholar
- Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. DOIPubMedGoogle Scholar
- Costa RL, Moreira J, Lorenzo A, Lamas CC. Infectious complications following probiotic ingestion: a potentially underestimated problem? A systematic review of reports and case series. BMC Complement Altern Med. 2018;18:329. DOIPubMedGoogle Scholar
- Linz B, Windsor HM, McGraw JJ, Hansen LM, Gajewski JP, Tomsho LP, et al. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat Commun. 2014;5:4165. DOIPubMedGoogle Scholar
- Hassoun-Kheir N, Snitser O, Hussein K, Rabino G, Eluk O, Warman S, et al. Concordance between epidemiological evaluation of probability of transmission and whole genome sequence relatedness among hospitalized patients acquiring Klebsiella pneumoniae carbapenemase-producing Klebsiella pneumoniae. Clin Microbiol Infect. 2021;27:468.e1–7. DOIPubMedGoogle Scholar
- Goldenberg JZ, Lytvyn L, Steurich J, Parkin P, Mahant S, Johnston BC. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev. 2015;12:
CD004827 . DOIPubMedGoogle Scholar - Goldenberg JZ, Ma SS, Saxton JD, Martzen MR, Vandvik PO, Thorlund K, et al. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev. 2013;5:
CD006095 . DOIGoogle Scholar - Xu J, Ma R, Chen LF, Zhao LJ, Chen K, Zhang RB. Effects of probiotic therapy on hepatic encephalopathy in patients with liver cirrhosis: an updated meta-analysis of six randomized controlled trials. Hepatobiliary Pancreat Dis Int. 2014;13:354–60. DOIPubMedGoogle Scholar
- Naidoo K, Gordon M, Fagbemi AO, Thomas AG, Akobeng AK. Probiotics for maintenance of remission in ulcerative colitis. Cochrane Database Syst Rev. 2011;12:
CD007443 . DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleOriginal Publication Date: February 27, 2024
Table of Contents – Volume 30, Number 4—April 2024
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Ryuichi Minoda Sada, Department of Transformative Protection to Infectious Disease, Graduate School of Medicine, Osaka University, Osaka, Japan
Top