Volume 30, Number 6—June 2024
Dispatch
SARS-CoV-2 in Captive Nonhuman Primates, Spain, 2020–2023
Cite This Article
Citation for Media
Abstract
We conducted a serologic and molecular study to assess exposure of captive nonhuman primates (NHPs) to SARS-CoV-2 in Spain during the 2020–2023 COVID-19 pandemic. We found limited exposure of NHPs to SARS-CoV-2. Biosafety measures must be strictly maintained to avoid SARS-CoV-2 reverse-zoonotic transmission in the human–NHP interface.
COVID-19 caused by SARS-CoV-2 is an emerging respiratory disease that likely originated from wildlife in late 2019 (1). Although the main driver of SARS-CoV-2 spread is human-to-human transmission, several wild and domestic mammals are susceptible to SARS-CoV-2 infection; natural infections have been reported in 29 nonhuman animal species (2).
Nonhuman primates (NHPs) are among the most susceptible taxonomic groups to SARS-CoV-2 infection (3) and are suitable animal models to evaluate SARS-CoV-2 pathogenesis (4). Natural infections with clinical outcomes have been reported in both captive and free-living NHPs worldwide (2,5–8). Because active surveillance of SARS-CoV-2 has not been conducted in NHPs, we assessed SARS-CoV-2 circulation among NHPs housed in zoos and rescue centers in Spain, where NHPs can be found in proximity to humans.
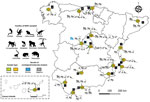
During January 2020–March 2023, we collected serum samples from 127 different NHPs belonging to 30 species housed in 17 zoos and NHP rescue centers in Spain. The number of sampled animals represented ≈40% of the total census of those species in the selected zoos and rescue centers. We collected 16 serum samples in 2020, 76 in 2021, 28 in 2022, and 7 in 2023 (Table; Figure 1). In addition, we collected 186 fresh fecal samples during February–May 2022 within the same zoos and NHPs rescue centers, which comprised 39 samples collected from animals housed in individual facilities and 147 pooled samples from the floors of facilities with >1 animal (1 pool per facility and species) belonging to 64 different NHP species (Appendix Table). The study did not require additional blood extractions because we obtained all serum samples from serum banks or from NHPs that had medical check-ups or surgical interventions during the study period.
We tested all serum samples for antibodies against SARS-CoV-2 nucleocapsid (N) and spike (S) proteins by using 2 commercial multispecies ELISAs (ID Screen SARS-CoV-2 Double Antigen and NeutraLISA SARS-CoV-2; Euroimmun, https://www.euroimmun.com) according to the manufacturer’s instructions. We analyzed ELISA-positive serum samples further by using a virus neutralization test (VNT) as previously described (9). In brief, we preincubated 100 SARS-CoV-2 (B.1 lineage) 50% tissue culture infectious doses with 2-fold serial dilutions (1:20 to 1:10,240) of heat-inactivated serum samples in Nunc 96-well cell culture plates (Thermo Fisher Scientific, https://www.thermofisher.com) for 30 minutes at 37°C. Then, we added the virus-serum mixtures onto Vero cells (ATCC, https://www.atcc.org) and read results after 72 hours by using the Cell Titer Glo kit (Promega, https://www.promega.com). We normalized values and calculated VNT50 (the reciprocal dilution inhibiting 50% of Vero cell infection) by plotting and fitting the log of serum dilution versus normalized response in Prism 8.4.3 (Graphpad, https://www.graphpad.com). We performed VNTs in duplicate for each sample.
We tested fecal samples for SARS-CoV-2 RNA by using quantitative reverse transcription PCR. We extracted RNA from feces by using the IndiSpin QIAcube HT Pathogen Kit (Indical Biosciences, https://www.indical.com) according to the manufacturer’s instructions. We detected SARS-CoV-2 RNA by using a previously published method (10) that had minor modifications adapting it to the Applied Biosystems AgPath-ID One-Step RT-PCR Kit (Thermo Fisher Scientific). We performed PCR amplification by using an Applied Biosystems 7500 Fast Real-Time PCR System (Thermo Fisher Scientific) and considered samples with a cycle threshold value of <40 to be positive for SARS-CoV-2 RNA.
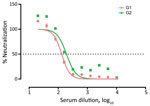
We confirmed SARS-CoV-2 antibodies in 2/127 (1.6% [95% CI 0.0%–3.7%]) tested animals by both ELISA and VNT. The seropositive animals were 2 western lowland gorillas (Gorilla gorilla gorilla) designated as G1 and G2. VNT50 titers were 1:131.4 for G1 and 1:191.9 for G2 serum samples (Figure 2).
G1 and G2 were adult female gorillas sampled at zoo D within the same enclosure (Table; Figure 1). G1 arrived at zoo D from a UK zoo on March 4, 2020, after a favorable medical evaluation. During April 7–10, zookeepers reported that G1 was exhibiting a dry cough. A serum sample was collected from G1 on April 24, 2020, during a clinical intervention after an aggression was suffered by another gorilla from the same group. Previous studies of NHPs experimentally infected with SARS-CoV-2 reported clinical signs within the first week after infection (4,11). Therefore, the exposure of G1 to SARS-CoV-2 most likely occurred during the first wave of the COVID-19 pandemic in Spain (February–June 2020), when the zoo was closed to the public because of lockdown efforts to curb coronavirus cases. G2 was sampled on November 4, 2022, and did not show previous clinical signs compatible with SARS-CoV-2 infection. The 4 group members that shared the same facilities with the 2 seropositive gorillas could not be sampled, but they did not show any indications of disease at that time.
Previous cases of natural SARS-CoV-2 infections have been described in gorillas; the animals experienced asymptomatic or mild illness, involving coughing, congestion, nasal discharge, loss of appetite, and tiredness that did not require medical interventions and resolved within a few days (5,6,8,12). In those previous cases, intraspecific transmission among animals from the same facilities was suggested.
Despite the rigorous biosafety protocols used when working with NHPs (e.g., using disinfection mats, masks, gloves, and hand disinfectants and changing overalls), the zoo staff was the most plausible source of transmission to the gorilla troop, which has been suggested for previous outbreaks reported in zoos (6,8). Nevertheless, exposure of G1 to SARS-CoV-2 at the zoo of origin or during transport, as well as intraspecific transmission, cannot be ruled out.
Fecal shedding of SARS-CoV-2 peaks during the symptomatic period and can persist 1–35 days after onset of clinical signs in humans (13,14). Similarly, a study of rhesus macaques (Macaca mulatta) experimentally infected with SARS-CoV-2 reported that rectal swab samples tested positive for RNA up to 27 days after infection (15). In our study, SARS-CoV-2 RNA was not detected in any of the 186 fecal samples tested (Appendix Table), including those obtained at the facility housing G1 and G2. Considering the time of fecal sampling and the virus excretion period in primate species (≈1 month), our findings indicate an absence of virus circulation during this temporal window. Discrepancies between serologic and molecular results in the 2 seropositive gorillas could be explained by differences in antibody persistence, which ranged from 3 to >9 months in experimentally immunized NHPs (Appendix references 16,17), and by differences in virus RNA excretion in feces, as well as sampling times (April 2020 and November 2022 for serum samples and February 2022 for fecal samples).
The first limitation of this study is that the spatial distribution of sampling was not homogeneous. Approximately 60% of the tested serum samples were from 4 of the sampled centers. Second, serum samples could not be longitudinally analyzed. Because antibodies decay over time (Appendix references 16,17), SARS-CoV-2 exposure before the sampling period cannot be ruled out. Further longitudinal studies would be of great interest to better understand the temporal dynamics of SARS-CoV-2 in NHPs.
Our findings indicate a limited SARS-CoV-2 exposure in captive NHPs populations in zoos and NHPs rescue centers in Spain during the 2020–2023 COVID-19 pandemic period. G1 only exhibited mild symptoms, and G2 exhibited no symptoms, highlighting the importance of conducting active screening and surveillance testing to reduce the potential emergence of unidentified reservoirs and virus evolution. Considering the potential health risk and threat to the conservation of NHP species, the possibility of the emergence of new reservoirs, and the opportunities for virus evolution, vaccination of captive animals and the proper use of biosafety measures and personal protective equipment are critical to prevent future reverse-zoonotic transmission of SARS-CoV-2.
Dr. Cano-Terriza is an assistant professor in the Department of Veterinary Medicine at the University of Cordoba. His research interests focus on the epidemiology of emerging and reemerging zoonotic diseases.
Acknowledgments
We thank Bioparc Fuengirola, Bioparc Valencia, Centro de Conservación Zoo Córdoba, Faunia, Marcelle Naturaleza, MundoMar Benidorm, Parque de la Naturaleza de Cabárceno, Selwo Aventura, Terra Natura Murcia, Zoo de Santillana del Mar, and Centro de Conservación de la Biodiversidad Zoobotánico Jerez-Alberto Durán for providing the valuable samples.
This research was partially supported by the Innovation and Transfer Plan of the University of Cordoba and CIBER, Consorcio Centro de Investigación Biomédica en Red (CB2021/13/00083), Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación and Unión Europea–NextGenerationEU. A.B.-B. is a PhD. candidate supported by the Andalusian System of Knowledge.
References
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. DOIPubMedGoogle Scholar
- World Organisation for Animal Health. SARS-CoV-2 in animals—situation report 22. 2023 [cited 2023 July 1] https://www.woah.org/app/uploads/2023/07/sars-cov-2-situation-report-22.pdf
- Melin AD, Janiak MC, Marrone F III, Arora PS, Higham JP. Comparative ACE2 variation and primate COVID-19 risk. Commun Biol. 2020;3:641. DOIPubMedGoogle Scholar
- Meekins DA, Gaudreault NN, Richt JA. Natural and experimental SARS-CoV-2 infection in domestic and wild animals. Viruses. 2021;13:1993. DOIPubMedGoogle Scholar
- Gibbons A. Captive gorillas test positive for coronavirus. Scienceinsider. January 12, 2021 [cited 2023 May 23]. https://www.science.org/content/article/captive-gorillas-test-positive-coronavirus
- Nagy A, Stará M, Vodička R, Černíková L, Jiřincová H, Křivda V, et al. Reverse-zoonotic transmission of SARS-CoV-2 lineage alpha (B.1.1.7) to great apes and exotic felids in a zoo in the Czech Republic. Arch Virol. 2022;167:1681–5. DOIPubMedGoogle Scholar
- Pereira AH, Vasconcelos AL, Silva VL, Nogueira BS, Silva AC, Pacheco RC, et al. Natural SARS-CoV-2 infection in a free-ranging black-tailed marmoset (Mico melanurus) from an urban area in mid-west Brazil. J Comp Pathol. 2022;194:22–7. DOIPubMedGoogle Scholar
- Dusseldorp F, Bruins-van-Sonsbeek LGR, Buskermolen M, Niphuis H, Dirven M, Whelan J, et al. SARS-CoV-2 in lions, gorillas and zookeepers in the Rotterdam Zoo, the Netherlands, a One Health investigation, November 2021. Euro Surveill. 2023;28:
2200741 . DOIPubMedGoogle Scholar - Fernández-Bastit L, Rodon J, Pradenas E, Marfil S, Trinité B, Parera M, et al. First detection of SARS-CoV-2 Delta (B.1.617.2) variant of concern in a dog with clinical signs in Spain. Viruses. 2021;13:2526. DOIPubMedGoogle Scholar
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:
2000045 . DOIPubMedGoogle Scholar - Yuan L, Tang Q, Zhu H, Guan Y, Cheng T, Xia N. SARS-CoV-2 infection and disease outcomes in non-human primate models: advances and implications. Emerg Microbes Infect. 2021;10:1881–9. DOIPubMedGoogle Scholar
- Zoo Atlanta. Update on gorilla population—Sept 17. September 17, 2021 [cited 2023 Jun 23]. https://zooatlanta.org/update-on-gorilla-population-sept-17
- Jones DL, Baluja MQ, Graham DW, Corbishley A, McDonald JE, Malham SK, et al. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci Total Environ. 2020;749:
141364 . DOIPubMedGoogle Scholar - Ma X, Su L, Zhang Y, Zhang X, Gai Z, Zhang Z. Do children need a longer time to shed SARS-CoV-2 in stool than adults? J Microbiol Immunol Infect. 2020;53:373–6. DOIPubMedGoogle Scholar
- Zheng H, Li H, Guo L, Liang Y, Li J, Wang X, et al. Virulence and pathogenesis of SARS-CoV-2 infection in rhesus macaques: A nonhuman primate model of COVID-19 progression. PLoS Pathog. 2020;16:
e1008949 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: May 14, 2024
Table of Contents – Volume 30, Number 6—June 2024
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
David Cano-Terriza, Departamento de Sanidad Animal, Campus universitario de Rabanales, Carretera Madrid Km 396, Universidad de Córdoba, Cordoba 14014, Spain
Top