Evidence of Orientia spp. Endemicity among Severe Infectious Disease Cohorts, Uganda
Paul W. Blair

, Kenneth Kobba, Stephen Okello, Sultanah Alharthi, Prossy Naluyima, Emily Clemens, Hannah Kibuuka, Danielle V. Clark, Francis Kakooza, Mohammed Lamorde, Yukari C. Manabe, J. Stephen Dumler, Acute Febrile Illness, and
Sepsis in Uganda study teams1
Author affiliations: Uniformed Services University, Bethesda, Maryland, USA (P.W. Blair, S. Alharthi, E. Clemens, J.S. Dumler); Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., Bethesda (P.W. Blair, S. Alharthi, D.V. Clark); Johns Hopkins University School of Medicine, Baltimore, Maryland, USA (P.W. Blair, Y.C. Manabe); Infectious Diseases Institute, Makerere University, Kampala, Uganda (K. Kobba, F. Kakooza, M. Lamorde); Makerere University Walter Reed Project, Kampala (S. Okello, P. Naluyima, H. Kibuuka)
Main Article
Figure 2
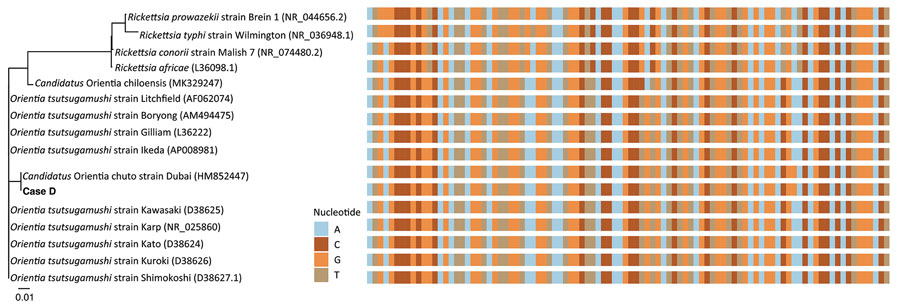
Figure 2. Phylogenetic tree (left) and aligned sequences (right) of Orientia spp. and locally endemic Rickettsia spp. in a study of Orientia genus endemicity among severe infectious disease cohorts, Uganda. We compared the 16S rRNA gene with an Orientia infection (case D) in Uganda. We aligned the 96-bp amplicon region and created the tree by using the neighbor-joining algorithm in R (The R Foundation for Statistical Computing, https://www.r-project.org). GenBank accession numbers of reference sequences are in parentheses. A single polymorphism aligned with Candidatus O. chuto, possibly differentiating case D from other Orientia spp. Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: May 10, 2024
Page updated: June 22, 2024
Page reviewed: June 22, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
