Volume 30, Number 7—July 2024
Dispatch
Evidence of Orientia spp. Endemicity among Severe Infectious Disease Cohorts, Uganda
Cite This Article
Citation for Media
Abstract
At 3 severe infection cohort sites in Uganda, Orientia seropositivity was common. We identified 4 seroconversion cases and 1 PCR-positive case. These results provide serologic and molecular support for Orientia spp. circulating in sub-Saharan Africa, possibly expanding its endemic range. Orientia infections could cause severe illness and hospitalizations in this region.
Scrub typhus is a leading cause of nonmalarial febrile illness in Southeast Asia (1). Scrub typhus is caused by miteborne Orentia tsutsugamushi infections, which until recently were thought to be limited to South and Southeast Asia. Molecular identification of different Orientia species in clinical cases from Chile (2) and the United Arab Emirates (3) has suggested a broader epidemiology. Orientia spp. were found in mites in Kenya (4), and descriptions of Orientia seroconversion in patients from sub-Saharan Africa have slowly accrued, suggesting the possibility of Orientia spp. transmission in Africa (5). We used archived samples collected in 2 severe infection prospective cohorts in western, central, and northwest Uganda to assess Orientia endemicity in the country.
Using archived samples, we measured serial Orientia immunofluorescence assay (IFA) IgG titers and performed reflex Orientia spp. reverse transcription PCR (RT-PCR). Samples were collected as part of 2 severe infection prospective cohorts and had undergone broad microbiologic testing. In both cohorts, adult patients >18 years of age who fulfilled acute febrile illness (AFI; hospitals in Mubende and Arua, Uganda) or sepsis-specific (hospital in Fort Portal, Uganda) eligibility criteria were evaluated for enrollment at admission in the outpatient or emergency department, or on medical wards (Appendix) (6). Matched acute and convalescent serum samples were available from 269 of 310 participants enrolled in the sepsis cohort and 67 of 132 participants in the AFI cohort.
In brief, across both prospective cohorts, study teams collected demographic and symptom information, examination findings, and laboratory data on standardized forms during hospitalization and at 1 month after enrollment. Clinical tests were routinely performed, including complete blood counts and chemistries. Microbiologic testing included blood culture with antimicrobial sensitivity testing, HIV testing, malaria smears, and rapid diagnostic tests, as previously described (6) (Appendix).
We performed IgG IFAs by using Orientia tsutsugamushi Karp strain antigen slides (BIOCELL Diagnostics Inc., https://biocelldx.com). Baseline (acute) and 1-month follow-up (convalescent) serum samples were screened at a titer of 1:64 and titrated up to 1:65,000. We considered a sample seropositive at a threshold titer of >128. We performed IgG IFAs by using commercial slides to evaluate for cross-reactivity to spotted fever group rickettsia (SFGR), Rickettsia conorii Molish 7 strain, typhus group rickettsia (TGR), and Rickettsia typhi Wilmington strain (BIOCELL Diagnostics, Inc.). We performed a Kruskal-Wallis test to evaluate for differences between Orientia IFA IgG titers between those with and without available matched samples. We used a titer of 32 to calculate -fold increase if the screen was negative at a titer of 1:64. We had a blind second reader review <5% of each batch.
Because no prior estimates of Orientia seroprevalence were available for Uganda, we used stringent criteria to define probable cases (Appendix Figure 1). To evaluate the specificity of IFA results, we used a subset of high titer samples to corroborate evidence of antibody binding by using a dot blot, Western blot, and Gilliam strain IFA (Appendix Methods, Figure 2). To optimize sensitivity for RT-PCR, we targeted mRNA and rRNA from serum from both cohorts (7), whole blood from the AFI cohort, or buffy coat from the sepsis cohort. We used QIAamp RNA Mini Kit (QIAGEN, https://www.qiagen.com) to extract RNA. We performed RT-PCR targeting Orientia spp. 16S rRNA, Orien16S and rrs by using previously published methods (3,8), and mRNA from Orientia spp. 56-kDa antigen gene, SFGR OmpA (sca0) gene, and TGR kDa (9) outer membrane protein gene. We only called positives that were in duplicate.
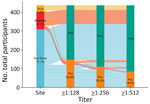
We found that 33.9% (148/436) of acute samples and 38.4% (129/336) of convalescent samples were seropositive (>128) for Orientia spp. Among acute samples, 25.5% (111/436) were positive at >256 titer and 19.0% (85/436) were positive at ≥512 (Figure 1). We observed no difference in acute IFA titers between patients with and without a convalescent blood samples (p = 0.33). Among samples with a positive 1:64 titer screen, the median acute titer was 128 (up to 8,192; interquartile range [IQR] 64–512) and median convalescent titer was 256 (up to 4,096; IQR 64–1,024). Seropositivity was highest (acute, 38.7% [120/310]; convalescent, 41.6% [112/269]) in Fort Portal, but was also high in Arua (acute, 26.5% [9/34]; convalescent, 30.0% [6/20]) and Mubende (acute, 20.7% [19/92]; convalescent, 23.4% [11/47]). The overall geometric mean titers were 90.8 (95% CI 80.2–102.8) for acute samples and 100.3 (95% CI 86.1–116.9) for convalescent samples.
Four participants met our case definition for Orientia spp. seroconversion (Table 1). Participants meeting the case definition were 24–56 years of age; 3 were female and 1 was male, and 3 had HIV (Table 2). Leukocyte counts ranged from 5–10 × 103 cells/μL, platelet counts were 56–220 × 103 cells/μL, and aspartate transaminase was 21–136 U/L. Three patients survived, but a 34-year-old woman with HIV in whom a papular rash developed died of unknown causes 8 months after follow-up. Three participants with seroconversion had negative malaria smears, blood cultures, and rapid antigen and molecular diagnostic tests for nonrickettsial pathogens (Table 2).
We used molecular methods to confirm Orientia spp. infection. The acute serum sample from participant D was repeatedly rrs-positive with RT-PCR (mean cycle threshold 34.1, SD 0.4) and was confirmed by Sanger sequencing of the amplicon. A BLAST analysis (https://blast.ncbi.nlm.nih.gov) of a 96-bp sequenced fragment of the amplicon revealed 96%–100% homology with Orientia spp., and a single polymorphism aligned with Candidatus O. chuto (Figure 2). RT-PCR was negative using other primers for Orientia spp. (Orien16S 56-kDa) targets, SFGR (sca0 [ompA] targets, and TGR (17-kDa antigen gene) targets.
We identified Orientia seroconversion among 4 participants hospitalized with severe infection in sub-Saharan Africa. We demonstrated that Orientia seropositivity was common among patients admitted for severe infection at 3 hospitals in Uganda. Our findings of highly prevalent seropositivity at 3 sites, identification of seroconversion, and molecular confirmation of a case with otherwise negative broad microbiologic testing support Orientia circulation and raise suspicion for infections extending to East Africa.
Prior clinical evidence of suspected scrub typhus in Africa relied on case reports of returning travelers with Orientia seroconversion (5). In addition to seroconversion identified in this study, seroconversions were observed in a pediatric cohort in Kenya (3.6%; n = 10) (10), and in 1 case among 49 abattoir workers in Djibouti (11). Our well-characterized multisite results supplement the limited literature suggesting Orientia spp. infections in sub-Saharan Africa.
In addition to prior suggestive evidence, our results build on a shift in understanding of worldwide Orientia spp. clinical infections. SFGR and TGR test results were negative in our cohorts, decreasing the likelihood of cross-reactivity. Despite IFA being the preferred method for rickettsial diagnosis, intrinsic interobserver variability limitations exist (12); we aimed to reduce those limitations through our reading approach and seroconversion criteria. Although we were able to confirm an infection by using real-time RT-PCR, sequence results were limited to a small fragment of the abundant 16S rRNA. The clinical relevance requires further confirmation with Orientia culture growth and extended genome sequencing. Because we relied on convalescent serology, we might have missed early fatal cases, which could skew our results toward less severe illness. Research efforts are needed to characterize the circulating species, incidence, pathogenic potential, and clinical relevance of Orientia infections in East Africa.
In summary, our findings suggest Orientia spp. circulation within the human–environment interface in Uganda and suggest novel Orientia infections within severe infection cohorts in Uganda. After excluding common causes of infections, our findings provide evidence of locally acquired Orientia infections among adults in sub-Saharan Africa.
Dr. Blair is an infectious diseases physician-scientist at Uniformed Services University, Bethesda, Maryland, USA. His research interests include molecular and imaging approaches to clinically detect emerging infectious diseases.
Acknowledgments
Acute Febrile Illness and Sepsis in Uganda Study Team members: Nehkonti Adams, Rodgers R. Ayebare, Helen Badu, Melissa Gregory, Francis Kakooza, Mubaraka Kayiira, Willy Kayondo, Stacy M. Kemigisha Hannah Kibuuka, Abraham Khandathil, Prossy Naluyima, Edgar C. Ndawula, David F. Olebo, Matthew Robinson, Abdullah Wailagala, and Peter Waitt.
This study was conducted in compliance with the Declaration of Helsinki and Good Clinical Practice Guidelines. All participants signed written informed consent prior to study procedures. The investigators have adhered to the policies for protection of human subjects as prescribed in 45 CFR 46. Parent acute febrile illness cohort: The study and informed consent process were reviewed and approved by the Joint Clinical Research Centre (JCRC) Research Ethics Committee (JC1518) and the Uganda National Council for Science and Technology (UNCST), HS 371ES, and Johns Hopkins University School of Medicine Internal Review Board (IRB no. 00176961). Parent sepsis cohort: This protocol and informed consent were approved by the US Army Medical Research and Development Command Institutional Review (approval no. M-10573) and Makerere University School of Public Health (IRB no. 490). Secondary use protocol: this laboratory work was reviewed and received an exempt determination by Uniformed Services University (IRB no. DBS.2020.174).
Pathogen testing was supported by cooperative agreement with the Naval Medical Logistics Command (NMLC; agreement no. N626451920001) and by the Congressionally Directed Medical Research Program (agreement no. W81XWH-19-2-0057).
The opinions and assertions expressed herein are those of the authors and do not reflect the official policy or position of the Uniformed Services University of the Health Sciences, US Department of Defense, Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., or any other government or agency. Mention of trade names, commercial products, or organizations does not imply endorsement by the US government. Some of the authors of this work are employees of the US government. This work was prepared as part of their official duties. Title 17 U.S.C. x105 provides that “Copyright protection under this title is not available for any work of the United States government.” Title 17 U.S.C. x101 defines a US government work as a work prepared by a military service member or employee of the US government as part of that person’s official duties.
Y.C.M. receives research funding from Becton Dickinson, Quanterix, and Hologic, and receives funding support from miDiagnostics to Johns Hopkins University. Y.C.M. receives research funding from Becton Dickinson, Quanterix, and Hologic, and receives funding support from miDiagnostics to Johns Hopkins University. M.L. receives research funding support from Pfizer Inc. to Infectious Diseases Institute.
References
- Bonell A, Lubell Y, Newton PN, Crump JA, Paris DH. Estimating the burden of scrub typhus: A systematic review. PLoS Negl Trop Dis. 2017;11:
e0005838 . DOIPubMedGoogle Scholar - Weitzel T, Dittrich S, López J, Phuklia W, Martinez-Valdebenito C, Velásquez K, et al. Endemic scrub typhus in South America. N Engl J Med. 2016;375:954–61. DOIPubMedGoogle Scholar
- Izzard L, Fuller A, Blacksell SD, Paris DH, Richards AL, Aukkanit N, et al. Isolation of a novel Orientia species (O. chuto sp. nov.) from a patient infected in Dubai. J Clin Microbiol. 2010;48:4404–9. DOIPubMedGoogle Scholar
- Masakhwe C, Linsuwanon P, Kimita G, Mutai B, Leepitakrat S, Yalwala S, et al. Identification and characterization of Orientia chuto in trombiculid chigger mites collected from wild rodents in Kenya. J Clin Microbiol. 2018;56:e01124–18. DOIPubMedGoogle Scholar
- Richards AL, Jiang J. Scrub typhus: historic perspective and current status of the worldwide presence of Orientia species. Trop Med Infect Dis. 2020;5:49. DOIPubMedGoogle Scholar
- Blair PW, Kobba K, Kakooza F, Robinson ML, Candia E, Mayito J, et al. Aetiology of hospitalized fever and risk of death at Arua and Mubende tertiary care hospitals in Uganda from August 2019 to August 2020. BMC Infect Dis. 2022;22:869. DOIPubMedGoogle Scholar
- Yun NR, Kim CM, Kim DY, Seo JW, Kim DM. Clinical usefulness of 16S ribosomal RNA real-time PCR for the diagnosis of scrub typhus. Sci Rep. 2021;11:14299. DOIPubMedGoogle Scholar
- Jiang J, Martínez-Valdebenito C, Weitzel T, Farris CM, Acosta-Jamett G, Abarca K, et al. Development of a new genus-specific quantitative real-time PCR assay for the diagnosis of scrub typhus in South America. Front Med (Lausanne). 2022;9:
831045 . DOIPubMedGoogle Scholar - Reller ME, Dumler JS. Optimization and evaluation of a multiplex quantitative PCR assay for detection of nucleic acids in human blood samples from patients with spotted fever rickettsiosis, typhus rickettsiosis, scrub typhus, monocytic ehrlichiosis, and granulocytic anaplasmosis. J Clin Microbiol. 2020;58:e01802–19. DOIPubMedGoogle Scholar
- Maina AN, Farris CM, Odhiambo A, Jiang J, Laktabai J, Armstrong J, et al. Q fever, scrub typhus, and rickettsial diseases in children, Kenya, 2011–2012. Emerg Infect Dis. 2016;22:883–6. DOIPubMedGoogle Scholar
- Horton KC, Jiang J, Maina A, Dueger E, Zayed A, Ahmed AA, et al. Evidence of rickettsia and Orientia infections among abattoir workers in Djibouti. Am J Trop Med Hyg. 2016;95:462–5. DOIPubMedGoogle Scholar
- Phetsouvanh R, Thojaikong T, Phoumin P, Sibounheuang B, Phommasone K, Chansamouth V, et al. Inter- and intra-operator variability in the reading of indirect immunofluorescence assays for the serological diagnosis of scrub typhus and murine typhus. Am J Trop Med Hyg. 2013;88:932–6. DOIPubMedGoogle Scholar
Figures
Tables
Cite This Article1Members of the Acute Febrile Illness and Sepsis in Uganda study teams are listed at the end of this article.
Table of Contents – Volume 30, Number 7—July 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Paul W. Blair, Uniformed Services University, 4301 Jones Bridge Rd, Bethesda, MD 20814, USA
Top