Volume 30, Number 8—August 2024
Dispatch
Rustrela Virus in Wild Mountain Lion (Puma concolor) with Staggering Disease, Colorado, USA
Figure 1
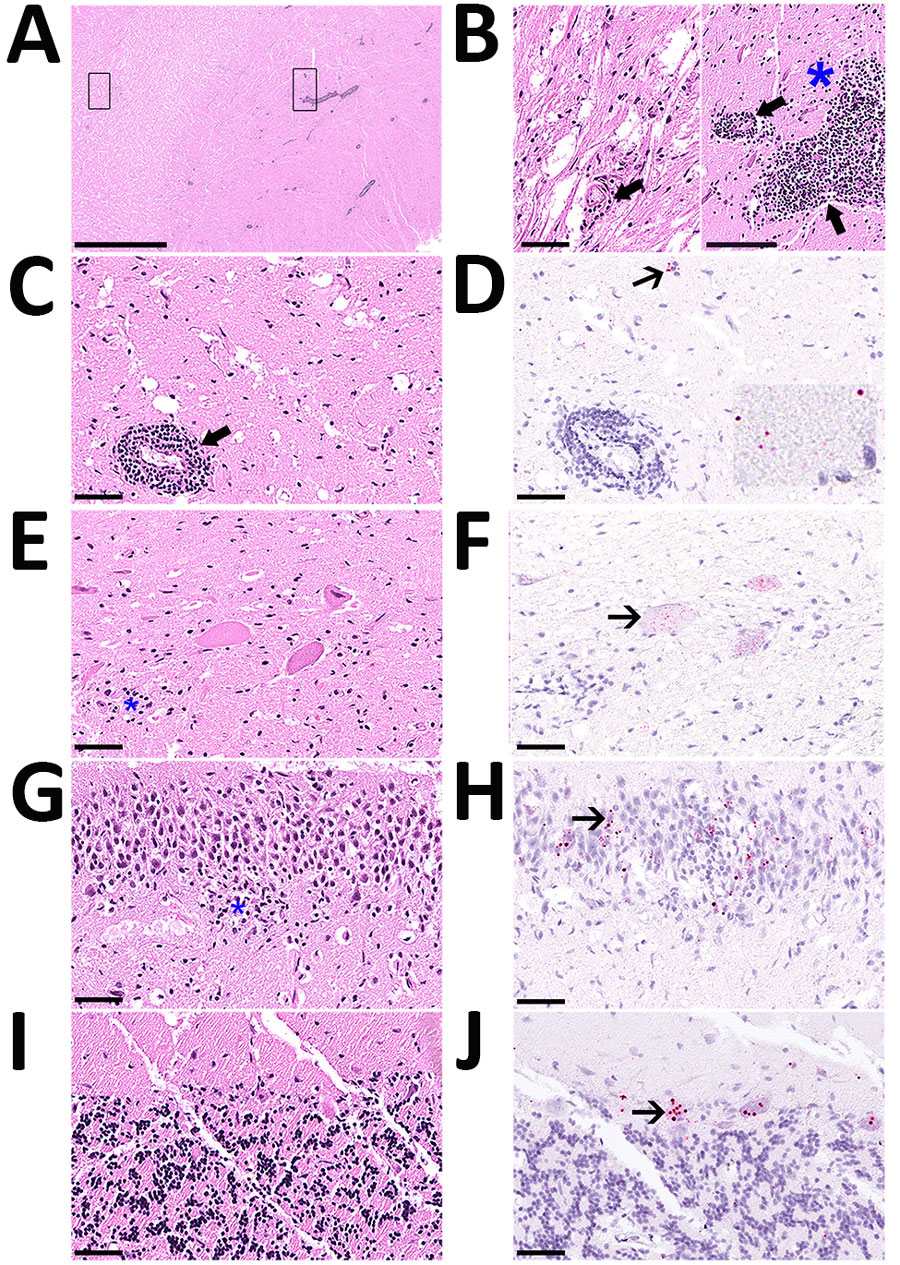
Figure 1. Histology of brain and spinal cord used to detect rustrela virus (RusV) in wild mountain lion (Puma concolor) with staggering disease, Colorado, USA. RusV RNA was detected by RNAscope Reagent Kit-Red (Advanced Cell Diagnostics/bio-techne, https://www.bio-techne.com) in situ hybridization. All sections demonstrate artifactual clefting of the neuropil due to freezing of the tissue postmortem. A) Cerebral cortex with perivascular cuffing and mild gliosis of the white and gray matter; boxes indicate detailed areas in panel B. Scale bar indicates 2.5 mm. B) The white matter (left panel) is minimally affected by perivascular lymphohistiocytic infiltrates (bold arrow), compared with the gray matter (right panel), also showing gliosis (asterisk). Scale bar indicates 100 µm. C) Midbrain affected by perivascular cuffing. Scale bar indicates 50 µm. D) Midbrain showing chromogenic labeling (fast red) of RusV in neuronal cell bodies (slender arrow) and in the neuropil (inlay). Scale bar indicates 50 µm. E) Spinal cord with 3 motor neurons showing variable degree of degeneration/necrosis and also gliosis (asterisk). Scale bar indicates 50 µm. F) Spinal cord with affected motor neurons with RusV RNA detection. Scale bar indicates 50 µm. G) Hippocampus exhibiting irregular architecture of the granule layer and gliosis (asterisk). Scale bar indicates 50 µm. H) Hippocampus with numerous RusV RNA signals in neurons of the granule cell layer in areas with or without irregular architecture. Scale bar indicates 50 µm. I) Cerebellum, no indication for inflammation or any degenerative process. Scale bar indicates 50 µm. J) Cerebellum with abundant RusV RNA labeling in Purkinje cells. Scale bar indicates 50 µm. A–C, E, G, I) Hematoxylin-eosin staining; D, F, H, I) RNAscope in situ hybridization with probes against the nonstructural protein–coding region of RusV, counterstained with Mayer’s hematoxylin.