Volume 30, Number 8—August 2024
Dispatch
Rustrela Virus in Wild Mountain Lion (Puma concolor) with Staggering Disease, Colorado, USA
Figure 2
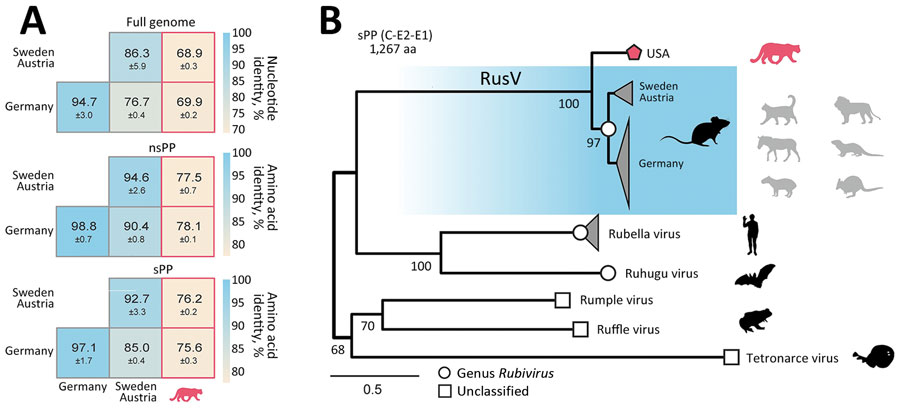
Figure 2. Sequence similarity and phylogenetic position of RusV in wild mountain lion (Puma concolor) with staggering disease, Colorado, USA. A) Mean pairwise sequence identity between the novel Colorado RusV and RusV sequences from Germany, Austria, and Sweden. Pairwise identity was based on nucleotide sequence alignments of the full genome or amino acid alignments of the nsPP and sPP. B) The sPP amino acid sequences of appropriate references from rubiviruses (circle) or currently unclassified matonavirids (square) were aligned with the novel RusV (pentagon). Phylogenetic tree was calculated using IQ-TREE (http://www.iqtree.org). Host species are depicted as silhouettes. For RusV, the potential reservoir (dark) and spillover hosts (light) are depicted. Scale bar indicates substitutions per site. nsPP, nonstructural polyprotein; sPP, structural polyprotein.