Volume 31, Number 10—October 2025
Dispatch
Fatal Pneumocephalus Caused by Hypervirulent Klebsiella pneumoniae, Germany
Figure 2
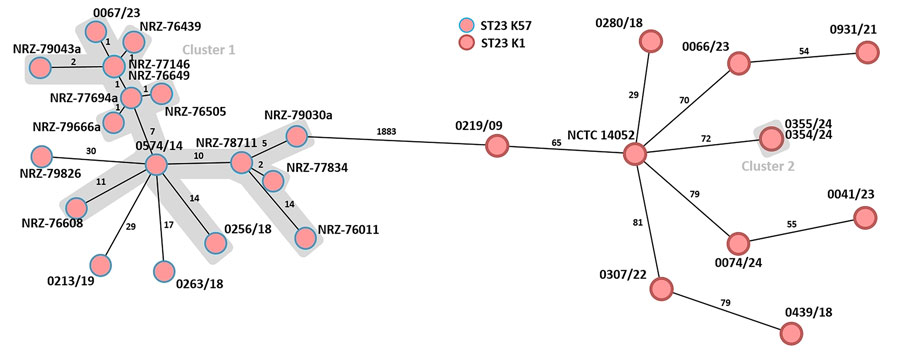
Figure 2. Minimum spanning tree generated to evaluate the genetic relationship of 29 Klebsiella pneumoniae-ST23 isolates (capsule types K1 and K57) during study of a fatal pneumocephalus case caused by hypervirulent K. pneumoniae in Germany (Appendix Table). Ridom SeqSphere version 10.0.4 (ridom, https://www.ridom.de) was used to create tree for 29 samples isolated from hospitals in Germany on the basis of 2,358 columns, pairwise ignoring missing values, and logarithmic scale. Distance is on the basis of columns from K. pneumoniae sensu lato core genome multilocus sequence type (2,358 genes). Cluster distance threshold is 15 according to the Ridom Seqsphere. The patient isolates from this case (0354/24 and 0355/24) are labeled as cluster 2. Isolate 0074/24 is another K. pneumoniae ST23-K1 isolate from a patient with liver abscess admitted to the same hospital. Cluster 1 represents K. pneumoniae ST23-K57 isolates with virulence or convergence plasmids that were in part from patients previously hospitalized in Ukraine (3,4). ST, sequence type.
References
- Das M. Global update on hypervirulent Klebsiella pneumoniae. Lancet Infect Dis. 2024;24:
e621 . DOIPubMedGoogle Scholar - Lee EJ, Kim RO, Lee M, Joo BE. Concurrent spontaneous pneumocephalus and subarachnoid hemorrhage due to Klebsiella pneumoniae meningitis. J Clin Neurol. 2022;18:253–5. DOIPubMedGoogle Scholar
- Wahl A, Fischer MA, Klaper K, Müller A, Borgmann S, Friesen J, et al. Presence of hypervirulence-associated determinants in Klebsiella pneumoniae from hospitalised patients in Germany. Int J Med Microbiol. 2024;314:
151601 . DOIPubMedGoogle Scholar - Sandfort M, Hans JB, Fischer MA, Reichert F, Cremanns M, Eisfeld J, et al. Increase in NDM-1 and NDM-1/OXA-48-producing Klebsiella pneumoniae in Germany associated with the war in Ukraine, 2022. Euro Surveill. 2022;27:
2200926 . DOIPubMedGoogle Scholar - Robertson J, Nash JHE. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom. 2018;4:
e000206 . DOIPubMedGoogle Scholar - Letunic I, Bork P. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–6. DOIPubMedGoogle Scholar
- Robertson J, Bessonov K, Schonfeld J, Nash JHE. Universal whole-sequence-based plasmid typing and its utility to prediction of host range and epidemiological surveillance. Microb Genom. 2020;6:mgen000435. DOIGoogle Scholar
- Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, Holt KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12:4188. DOIPubMedGoogle Scholar
- Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, et al. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom. 2016;2:
e000102 . DOIPubMedGoogle Scholar - Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. DOIPubMedGoogle Scholar
- Wu KM, Li LH, Yan JJ, Tsao N, Liao TL, Tsai HC, et al. Genome sequencing and comparative analysis of Klebsiella pneumoniae NTUH-K2044, a strain causing liver abscess and meningitis. J Bacteriol. 2009;191:4492–501. DOIPubMedGoogle Scholar
- Russo TA, Lebreton F, McGann PT. A step forward in hypervirulent Klebsiella pneumoniae diagnostics. Emerg Infect Dis. 2025;31:1–3. DOIPubMedGoogle Scholar
- Marr CM, Russo TA. Hypervirulent Klebsiella pneumoniae: a new public health threat. Expert Rev Anti Infect Ther. 2019;17:71–3. DOIPubMedGoogle Scholar
- Chang WN, Huang CR, Lu CH, Chien CC. Adult Klebsiella pneumoniae meningitis in Taiwan: an overview. Acta Neurol Taiwan. 2012;21:87–96.PubMedGoogle Scholar
- Neumann B, Stürhof C, Rath A, Kieninger B, Eger E, Müller JU, et al. Detection and characterization of putative hypervirulent Klebsiella pneumoniae isolates in microbiological diagnostics. Sci Rep. 2023;13:19025. DOIPubMedGoogle Scholar
1These first authors contributed equally to this article.