Volume 31, Number 11—November 2025
Dispatch
Novel Dolphin Tupavirus from Stranded Atlantic White-Sided Dolphin with Severe Encephalitis, Canada, 2024
Figure 2
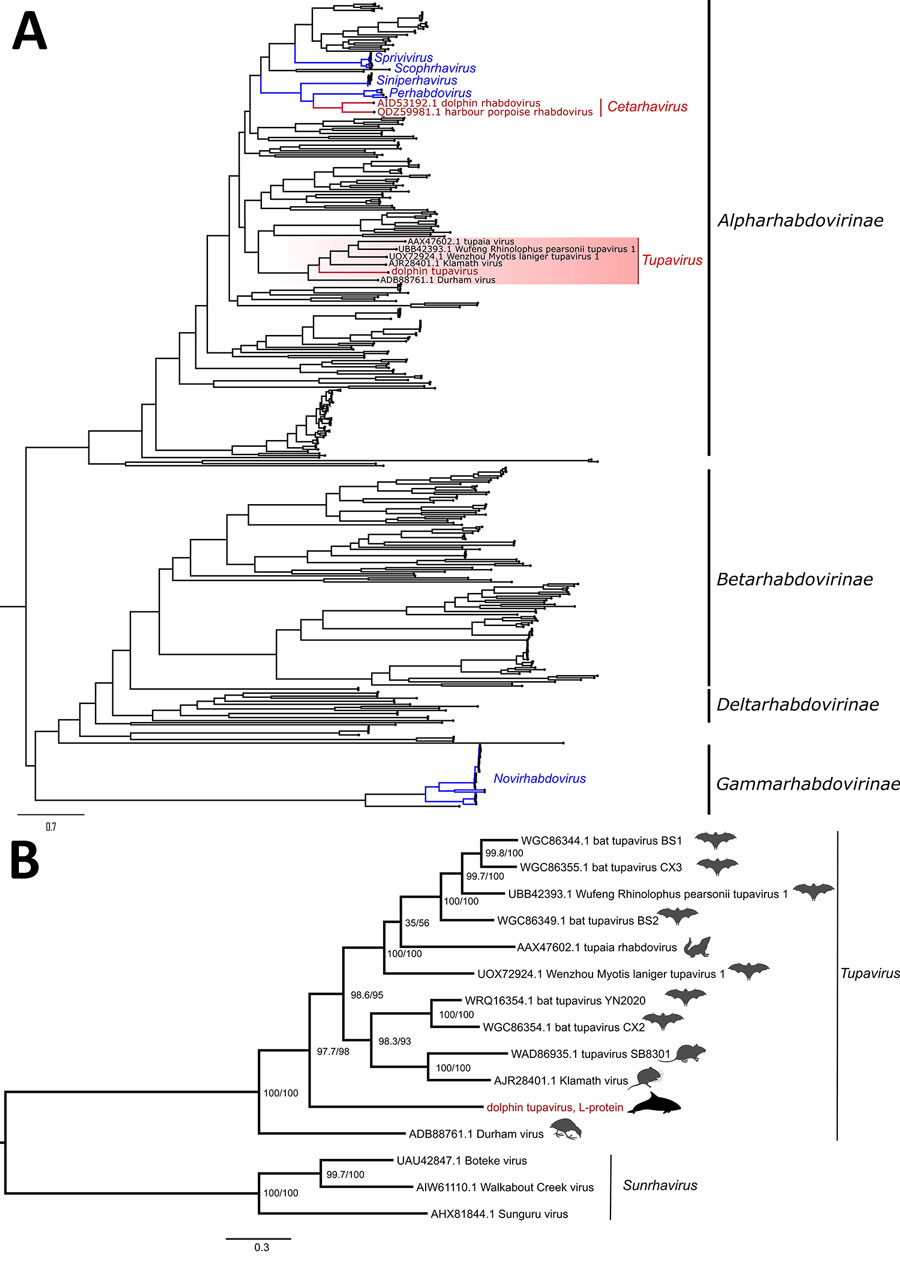
Figure 2. Maximum-likelihood phylogenetic trees of novel dolphin tupavirus from stranded Atlantic white-sided dolphin, Canada, and reference rhabdoviruses. Trees were reconstructed based on the full-length amino acid sequences of the polymerase protein. All sequence records for the phylogenetic analysis were downloaded from GenBank (Appendix 2 Table 2). Scale bars represent the estimated average number of substitutions per site. A) High-level phylogeny includes a subset of representative members across the Rhabdoviridae family (n = 438 sequence records) to determine the placement of the new virus within the family. Red text indicates rhabdoviruses of marine mammals, including the newly identified dolphin tupavirus; blue text indicates fish rhabdoviruses. The tree was rooted at the midpoint. B) Genus-level phylogeny provides a fine-level assessment of the phylogenetic affinities of the new virus within the Tupavirus genus. Red text indicates the sequence of the novel tupavirus; red shading highlights the Tupavirus clade in the phylogenetic tree. The phylogenetic bracket of the dolphin tupavirus includes a basal Durham virus and a large crown clade comprising the rest of the Tupavirus member viruses. Durham virus described from an American coot (Fulica americana) that demonstrated signs of severe disease associated with infection, including lesions isolated to the central nervous system and consisting of severe cerebral necrosis and mononuclear inflammation similar to DTV. Support values at the nodes were determined by SSH-aLRT/Ultrafast bootstrap. Tupavirus genus tree was rooted to the outgroup members of the Sunrhavirus genus, Boteke, Sunguru, and Walkabout Creek viruses.
1These authors contributed equally to this article.