Volume 31, Number 11—November 2025
Dispatch
Novel Dolphin Tupavirus from Stranded Atlantic White-Sided Dolphin with Severe Encephalitis, Canada, 2024
Abstract
We sequenced a novel rhabdovirus, Tupavirus delphini (dolphin tupavirus), from the brain of a stranded dead Atlantic white-sided dolphin with severe encephalitis in Canada. In situ hybridization linked presence of the virus to the animal’s brain pathology and death. Our findings underscore the importance of monitoring marine mammals for unexpected pathogens.
Cetaceans (whales and dolphins) are ubiquitous in the world’s oceans and are critical for monitoring oceanic ecosystem health (1). Despite their importance, little is known about diseases that affect free-ranging cetacean populations. Monitoring cetacean populations is challenging; consequently, much can be gained from necropsy examinations of dead, stranded animals. This study describes the discovery of a novel rhabdovirus species detected during the necropsy of an Atlantic white-sided dolphin (Leucopleurus acutus) found stranded on the Atlantic coast of Nova Scotia, Canada (Appendix 1 Figure 1).
Rhabdoviruses are a diverse group of enveloped single-stranded negative-sense RNA viruses that infect vertebrates, invertebrates, and plants. Most aquatic rhabdoviruses are described from ray-finned fish and amphibians; little is known about rhabdoviruses infecting marine mammals (2). Two species of rhabdoviruses are reported from cetaceans, dolphin rhabdovirus (3,4) and harbor porpoise rhabdovirus, neither of which was definitively linked to causing disease in their hosts (5). We characterized a novel rhabdovirus genome and used in situ hybridization to link viral infection with histopathologic lesions within the brain.
On October 27, 2024, a freshly dead Atlantic white-sided dolphin was found ashore on La Bloc Beach in the Cape Breton Highlands National Park, Nova Scotia, Canada. The carcass was well preserved with minimal scavenging or postmortem decomposition (Appendix 1 Figure 1) and demonstrated overall good nutritional condition (6). No significant lesions were identified from the carcass other than those from severe encephalitis. We conducted diagnostic PCR testing of frozen brain tissue for common pathogens known to cause encephalitis in marine mammals (avian influenza virus, cetacean morbillivirus, herpesvirus, Brucella ceti, Sarcocystis sp., and Toxoplasma gondii); results were negative.
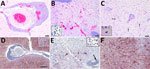
Examination of transverse sections of the brain revealed multiple extensive areas of severe inflammation and necrosis primarily throughout the forebrain within the regions of the cingulate gyrus, internal capsule, thalamus, and temporal and parietal lobes of the neocortex. In situ hybridization showed consistent presence of viral RNA in the lesions (Figure 1). Regions of the brain that did not exhibit lesions were primarily within the midbrain and hindbrain. In affected regions, we observed generalized mild to severe expansion of the perivascular space by lymphocytes and plasma cells and fewer macrophages and rare eosinophils (Figure 1, panel A). We observed intense positive staining for viral RNA in the surrounding neuropil but not in the perivascular inflammatory cells (Figure 1, panel D). In some areas, the normal tissue architecture was effaced by massive infiltration of hypertrophied microglial cells (Figure 1, panel B), many of which stained positive for viral RNA (Figure 1, panel E). In the neocortex, we noted glial nodules surrounding necrotic neurons (Figure 1, panel C), as well as neuronal cell bodies and dendrites within lesion areas that contained abundant viral RNA (Figure 1, panel F). All other tissues examined microscopically were either within normal limits or exhibited incidental lesions unrelated to the neuropathology described.
For whole-genome sequencing, we performed sample preparation and data processing (7) with modifications (Appendix 1). The complete 11,088-nt dolphin tupavirus (DTV) genome we obtained (deposited into GenBank, accession no. PV683224) had a structure typical of rhabdoviruses comprising 5 major open reading frames (ORFs), nucleocapsid, phosphoprotein, matrix, glycoprotein, and RNA-dependent RNA polymerase (L) proteins, and a putative small hydrophobic protein between the matrix protein and glycoprotein, that encode proteins highly divergent from other rhabdoviruses (Table; Figure 2) (Appendix 1 Figure 2; Appendix 2 Table 1) (8,9). Phosphoprotein contains a putative C protein in an overlapping reading frame. Each ORF of the DTV genome is flanked by conserved transcription initiation (UUGUC) and termination/polyadenylation (AWCU7) signals and an inferred untranscribed intergenic sequence (GG or GA). The L protein of DTV contains an LDSPL motif, a modification of the animal rhabdovirus conserved motif (LNSPL), also found in Durham virus, another tupavirus (10,11).
Phylogenetic analysis of the complete L-protein sequences placed DTV within the genus Tupavirus (Figure 2). The closest BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) match to the assembled DTV genome was Wenzhou Myotis laniger tupavirus 1 (GenBank accession no. OM030290.1), with an overall genomewide nucleotide identity of 50.72%. Amino acid sequence divergence in the nucleocapsid protein between the DTV and the closest BLAST match (Wufeng bat tupavirus 2; accession no. OQ715690.1) was 45.12%. Amino acid sequence divergence between the DTV and the closest BLAST match was 69.38% (Wufeng bat tupavirus 2; accession no. OQ715690.1) in the G protein and 44.64% (Klamath virus; accession no. KM204999.1) in the L protein. Virus isolation with multiple passages for >1 month using 6 cell lines from diverse host species was unsuccessful, as indicated by the lack of visible cytopathic effect and negative results with a DTV PCR (Appendix 2 Table 3).
We report the identification and characterization of a novel rhabdovirus from the brain of a stranded Atlantic white-sided dolphin with severe encephalitis. DTV is genetically distinct from previously known cetacean rhabdoviruses, and phylogenetic analysis indicates a considerable gap between DTV and other tupaviruses, suggesting a substantial level of unsampled viral diversity within the group. Based on the presented genomic divergence, genome organization, phylogenetic placement, and host species, DTV complies with International Committee on Taxonomy of Viruses demarcation criteria as a new species within the genus Tupavirus (10).
Reports of rhabdoviruses in aquatic mammals are scarce. Their detection and discovery are hindered by numerous challenges presented by the environment, host biology, and postmortem decomposition; therefore, even single case reports of a new virus from an aquatic mammal host provide valuable information to enhance pathogen surveillance in aquatic environments.
Unlike the previous 2 cases of cetacean rhabdovirus infection, DTV was associated with signs of severe disease. The severity of encephalitis we described was striking, but not unique. Many pathogens, including morbillivirus and Brucella ceti bacteria, can cause encephalitis in marine mammals (12). Those agents and others were ruled out in this case through a series of PCR tests, further supporting DTV as the causative pathogen. Even so, we cannot totally rule out the presence of concurrent infections. DTV isolation in cultured cells was challenging and unsuccessful; however, it is critical for experimental infections in animal models that will support investigations into DTV’s host range, tissue tropism, transmission, and pathogenicity. PCR screening and whole-genome sequencing methods for DTV could enable DTV surveillance. In addition, metagenomic sequencing of both affected and unaffected tissues might identify any co-infections.
The apparent preference of DTV for specific areas of the brain is reminiscent of rabies infection of terrestrial mammals, in which different host species exhibit unique patterns of lesion distribution in the brain; those patterns are an important criterion for disease recognition and diagnosis (13). A large outbreak of rabies has been reported in cape fur seals (Arctocephalus pusillus) in South Africa, which likely originated as a spillover event from terrestrial canids (14). Before that event, rabies in marine mammals was considered vanishingly rare; 1 case had been reported in a ring seal (Pusa hispida) in Svalbard, Norway, in 1981 (15).
This work highlights the importance of responding to incidents involving dead marine animals and conducting thorough investigations and diagnostics. Such discoveries can compound conservation concerns for marine species. It is imperative to continue documenting and examining cetacean stranding incidents in Canada, including those involving species not considered to be a priority by federal government wildlife managers. Our work has provided histopathologic and molecular evidence linking a cetacean rhabdovirus to CNS pathology and supports further investigation to characterize DTV and its associated pathology and epidemiology.
Dr. Vernygora is a research scientist at the National Centre for Foreign Animal Disease, Canadian Food Inspection Agency. Her research focuses on molecular identification and characterization of known, unknown, and unexpected animal viruses. Dr. Bourque is a wildlife pathologist that specializes in marine mammals. She works for the Canadian Wildlife Health Cooperative in Prince Edward Island, Canada.
Acknowledgments
We thank Jordi Segers for creating the map in Appendix 1 Figure 1. We thank the Animal Health Center (Abbotsford, BC) for PCR testing of suspected pathogens including avian influenza virus, cetacean morbillivirus, herpesvirus, Brucella ceti, Sarcocystis sp., and Toxoplasma gondii. We thank Parks Canada, Nova Scotia’s Department of Natural Resources, and Marine Animal Response Society volunteers for the collection of the dolphin carcass. We acknowledge Sharon Clouthier for providing SSN-1 cells and Brad Pickering and Josip Rudar for review and suggestions on the manuscript.
The MARS response program is made possible through the support of funders including the Government of Canada and private donors. The Canadian Food Inspection Agency and the interdepartmental Genomics Research and Development Initiative Shared Priority Project, GenARCC, and the Canadian Safety and Security Program provided funding support for O.L.
References
- Bossart GD. Marine mammals as sentinel species for oceans and human health. Vet Pathol. 2011;48:676–90. DOIPubMedGoogle Scholar
- Walker PJ, Bigarré L, Kurath G, Dacheux L, Pallandre L. Revised taxonomy of rhabdoviruses infecting fish and marine mammals. Animals (Basel). 2022;12:1363. DOIPubMedGoogle Scholar
- Osterhaus ADME, Broeders HWJ, Teppema JS, Kuiken T, House JA, Vos HW, et al. Isolation of a virus with rhabdovirus morphology from a white-beaked dolphin (Lagenorhynchus albirostris). Arch Virol. 1993;133:189–93. DOIPubMedGoogle Scholar
- Siegers JY, van de Bildt MWG, van Elk CE, Schürch AC, Tordo N, Kuiken T, et al. Genetic relatedness of dolphin rhabdovirus with fish rhabdoviruses. Emerg Infect Dis. 2014;20:1081–2. DOIPubMedGoogle Scholar
- Emelianchik A, Rodrigues TCS, Subramaniam K, Nielsen O, Burek-Huntington KA, Rotstein D, et al. Characterization of a novel rhabdovirus isolated from a stranded harbour porpoise (Phocoena phocoena). Virus Res. 2019;273:
197742 . DOIPubMedGoogle Scholar - Geraci JR, Lounsbury VJ. Marine mammals ashore: a field guide for strandings. College Station (TX): Texas A&M University, Sea Grant College Program; 1993.
- Vernygora O, Sullivan D, Nielsen O, Huntington KB, Rouse N, Popov VL, et al. Senecavirus cetus a novel picornavirus isolated from cetaceans represents a major host switching to the marine environment. Npj Viruses. 2024;2:33. DOIGoogle Scholar
- Allison AB, Palacios G, Travassos da Rosa A, Popov VL, Lu L, Xiao SY, et al. Characterization of Durham virus, a novel rhabdovirus that encodes both a C and SH protein. Virus Res. 2011;155:112–22. DOIPubMedGoogle Scholar
- Walker PJ, Firth C, Widen SG, Blasdell KR, Guzman H, Wood TG, et al. Evolution of genome size and complexity in the rhabdoviridae. PLoS Pathog. 2015;11:
e1004664 . DOIPubMedGoogle Scholar - Walker PJ, Freitas-Astúa J, Bejerman N, Blasdell KR, Breyta R, Dietzgen RG, et al.; Ictv Report Consortium. ICTV virus taxonomy profile: Rhabdoviridae 2022. J Gen Virol. 2022;103:
001689 . DOIPubMedGoogle Scholar - Kuzmin IV, Wu X, Tordo N, Rupprecht CE. Complete genomes of Aravan, Khujand, Irkut and West Caucasian bat viruses, with special attention to the polymerase gene and non-coding regions. Virus Res. 2008;136:81–90. DOIPubMedGoogle Scholar
- Gulland FMD, Dierauf LA, Whitman KL, editors. CRC handbook of marine mammal medicine. 3rd edition. Boca Raton (FL): CRC Press; 2018.
- Maxie M. Jubb, Kennedy, and Palmer’s pathology of domestic animals. 6th edition. St. Louis (MO): Elsevier; 2016.
- Winkler MP, Parker S. Rabies in seals: visitors to Cape Town marine areas urged to be alert. J Travel Med. 2024;31:taae106. DOIGoogle Scholar
- Odegaard OA, Krogsrud J. Rabies in Svalbard: infection diagnosed in arctic fox, reindeer and seal. Vet Rec. 1981;109:141–2. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: November 20, 2025
1These authors contributed equally to this article.
Table of Contents – Volume 31, Number 11—November 2025
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Oliver Lung, National Centre for Foreign Animal Disease, Canadian Food Inspection Agency, 1015 Arlington St, Winnipeg, MB R3E 3M4, Canada
Top