Novel Highly Pathogenic Avian Influenza A(H5N1) Virus, Argentina, 2025
Ralph E.T. Vanstreels, Martha I. Nelson, María C. Artuso, Vanina D. Marchione, Luana E. Piccini, Estefania Benedetti, Alvin Crespo-Bellido, Agostina Pierdomenico, Thorsten Wolff, Marcela M. Uhart, and Agustina Rimondi

Author affiliation: University of California Davis School of Veterinary Medicine, Davis, California, USA (R.E.T. Vanstreels, M.M. Uhart); National Institutes of Health, Bethesda, Maryland, USA (M.I. Nelson, A. Crespo-Bellido); Servicio Nacional de Sanidad y Calidad Agroalimentaria, Martínez, Buenos Aires, Argentina (M.C. Artuso, V.D. Marchione, L.E. Piccini); Instituto Nacional de Enfermedades Infecciosas Dr. Carlos G. Malbrán, Buenos Aires (E. Benedetti); Servicio Nacional de Sanidad y Calidad Agroalimentaria, Buenos Aires (A. Pierdomenico); Robert Koch-Institut, Berlin, Germany (T. Wolff, A. Rimondi)
Main Article
Figure 3
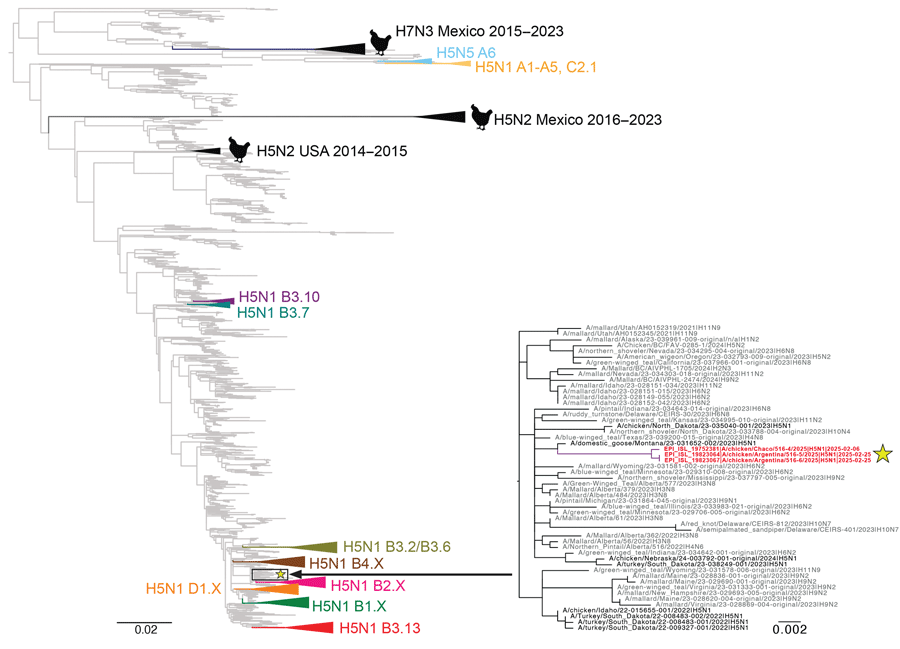
Figure 3. Phylogenetic tree showing how North American low pathogenicity avian influenza (LPAI) lineage contributed nucleoprotein (NP) genes by reassortment to novel influenza A(H5N1) viruses from Argentina, 2025. We inferred the phylogenetic tree by using the maximum-likelihood method for 11,820 North American LPAI and highly pathogenic avian influenza (HPAI) NP sequences collected during 2015–2025. Gray indicates LPAI viruses. HPAI H5N1 clades are collapsed and shaded in different colors and labeled according to corresponding H5 clade 2.3.4.4b genotypes. Black indicates prior H5N2 and H7N3 outbreaks in poultry. The 3 H5N1 viruses collected from poultry in Argentina in February 2025 are indicated in red and with a yellow star. A more detailed subsection of the tree containing those 3 viruses is shown, with tip labels. Branch lengths are drawn to scale. Scale bar represents nucleotide substitutions per site.
Main Article
Page created: November 17, 2025
Page updated: January 01, 2026
Page reviewed: January 01, 2026
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
