Bat Reovirus as Cause of Acute Respiratory Disease and Encephalitis in Humans, Bangladesh, 2022–2023
Sharmin Sultana
1, Ariful Islam
1, James Ng, Sunil Kumar Dubey, Manjur Hossain Khan, Cheng Guo, Mohammed Ziaur Rahman, Joel M. Montgomery, Syed Moinuddin Satter, Tahmina Shirin, W. Ian Lipkin, Lisa Hensley
2, and Nischay Mishra
2
Author affiliation: Institute of Epidemiology, Disease Control and Research (IEDCR), Dhaka, Bangladesh (S. Sultana, A. Islam, M. Hossain Khan, T. Shirin); Gulbali Research Institute, Charles Sturt University, Wagga Wagga, New South Wales, Australia (A. Islam); Center for Infection and Immunity, Mailman School of Public Health, Columbia University, New York, New York, USA (J. Ng, S. Kumar Dubey, C. Guo, W.I. Lipkin, N. Mishra); icddr,b, Dhaka (M.Z. Rahman, S. Moinuddin Satter); Centers for Disease Control and Prevention, Atlanta, Georgia, USA (J.M. Montgomery); Zoonotic and Emerging Disease Research Unit, National Bio and Agro-Defense Facility, USDA Agricultural Research Service, Manhattan, Kansas, USA (L. Hensley)
Main Article
Figure 3
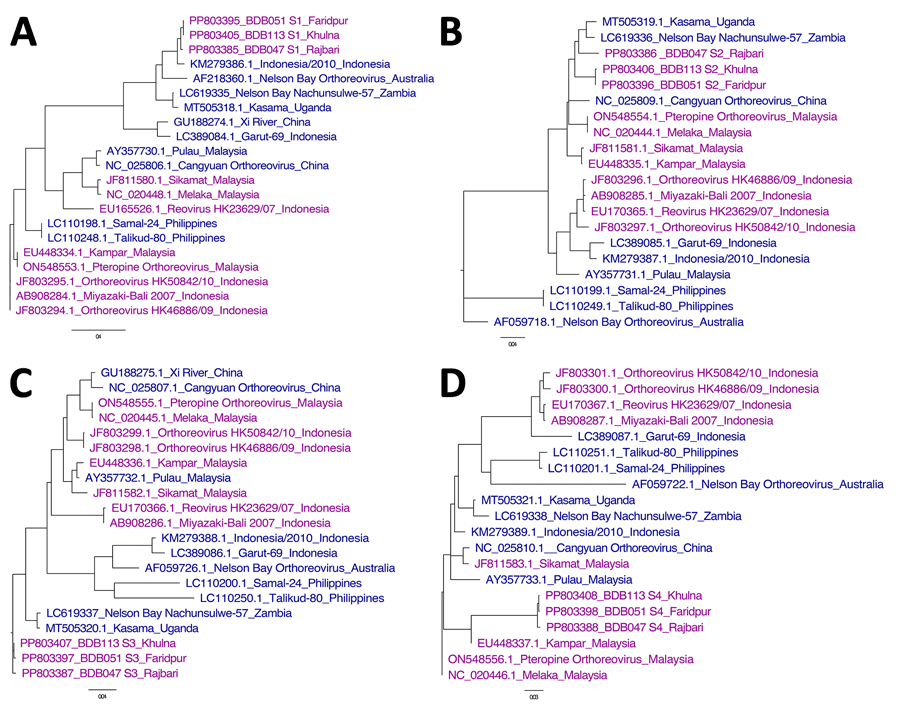
Figure 3. Comparitive phylogeny of bat reovirus as cause of acute respiratory disease and encephalitis in humans, Bangladesh, 2022–2023. A) S1 segment phylogeny showing 96.7%–99.9% average nucleotide identity (ANI) with each other (BDB051, BDB113, and BDB047) and clustered with Indonesia/2010, NBV-Australia, NBV-Nachunsulwe-57, and Kasama virus. B, C) Phylogeny of S2 (B) and S3 (C) segments showing partial consistency with S1 segments. D) S4 segments clustered with Kampar and Melaka strains, previously linked to mild respiratory illness in humans and reported human-to-human transmission. GenBank accession numbers are indicated. Scale bars indicate nucleotide substitutions per site.
Main Article
Page created: November 24, 2025
Page updated: January 01, 2026
Page reviewed: January 01, 2026
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
