Volume 31, Supplement—May 2025
SUPPLEMENT ISSUE
Supplement
Genomic Characterization of Escherichia coli O157:H7 Associated with Multiple Sources, United States
Figure 2
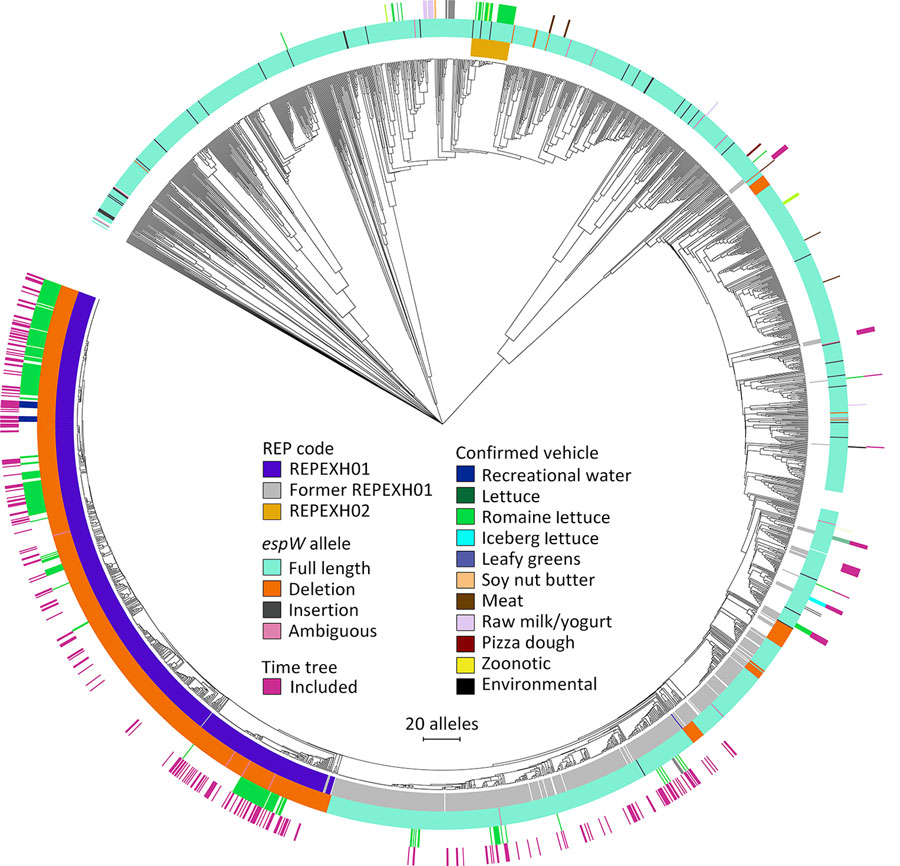
Figure 2. Allele-based core genome multilocus sequence typing dendrogram for 2,752 STEC O157:H7 isolates and associated metadata used for genomic characterization of Escherichia coli O157:H7 associated with multiple sources, United States. The dendrogram was constructed in BioNumerics version 7.6.3 (Applied Maths, http://www.applied-maths.com) by using UPGMA as the clustering technique. The innermost track indicates whether an isolate was included in a STEC O157:H7 REP strain. The second innermost track indicates the espW allele showing the full-length allele, single base pair deletions or insertion, where espW allele could not be determined due to inadequate sequencing data, and where espW was not detected in the isolate. The second outermost track indicates vehicles that have been confirmed via epidemiologic investigations of coded outbreaks. The outermost track indicates whether an isolate was included in the time-calibrated tree (Figure 1). An interactive version of this tree is available at https://itol.embl.de/tree/1581112362321361691692003. The following isolates were omitted from this tree: National Center for Biotechnology Information Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra) accession nos. SRR7094189, SRR8956189, SRR7540755, and SRR5588761. The scale bar indicates the number of allele differences between isolates. REP, reocurring, emerging, and persistent; REPHEX01, recurring strain of STEC HO1:H7; STEC, Shiga toxin–producing Escherichia coli.