Volume 31, Number 2—February 2025
Dispatch
Eastern Africa Origin of SAT2 Topotype XIV Foot-and-Mouth Disease Virus Outbreaks, Western Asia, 2023
Figure 3
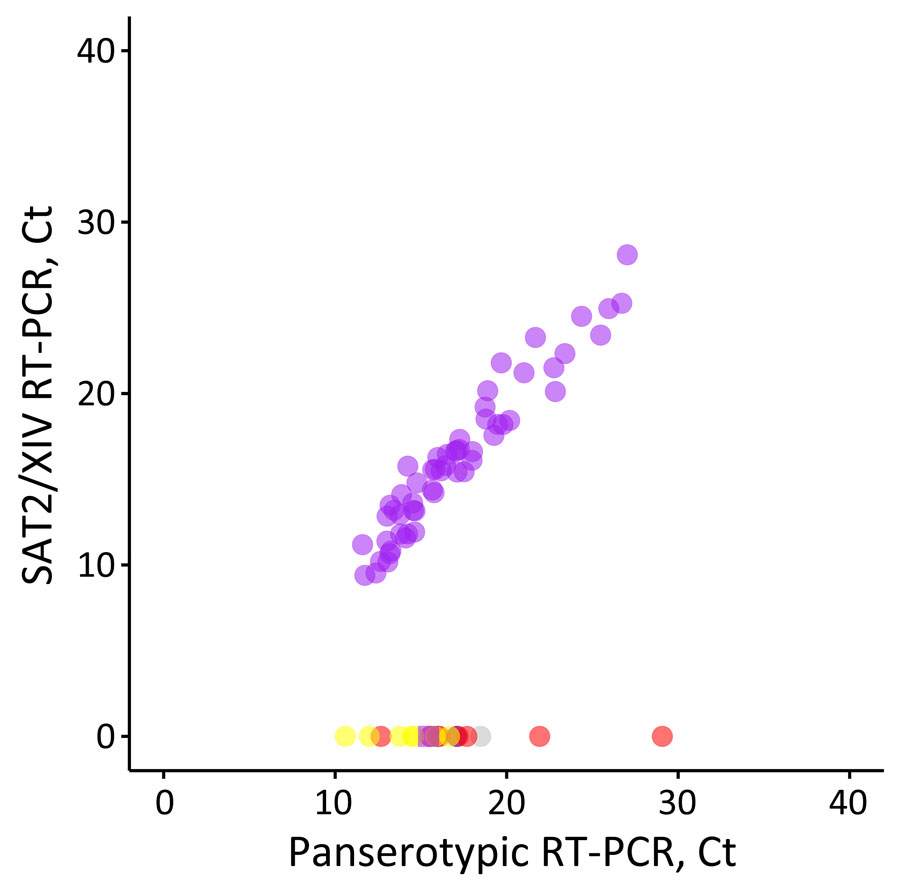
Figure 3. Performance of the SAT2/XIV-specific real-time RT-PCR for foot-and-mouth disease virus–positive samples representing different viral lineages and topotypes that are circulating or threaten countries in western Asia in study of eastern Africa origin of SAT2 topotype XIV foot-and-mouth disease virus outbreaks, western Asia, 2023. Plotted data shows real-time RT-PCR results for the SAT2/XIV specific test compared with results for the pan-serotype (3D) assay performed in parallel (5). Samples tested included clinical samples collected from confirmed SAT2/XIV cases (purple, n = 55) and characterized isolates from the O/ME-SA/PanAsia-2, O/ME-SA/Ind-2201 and O/EA-3 lineages (red, n = 8), the A/ASIA/Iran-05 lineage (blue, n = 1), the SAT1/I topotype (yellow, n = 7), the SAT2/VII topotype (purple, n = 8), and serotype Asia 1 (gray, n = 1). Ct, cycle threshold; RT-PCR, reverse transcription PCR.
References
- The Pirbright Institute. FAO World Reference Laboratory for foot-and-mouth disease genotyping report, Iraq (WRLMEG-2023-0004) [cited 2024 Feb 26]. https://www.wrlfmd.org/sites/world/files/quick_media/WRLMEG-2023-00004-IRQ-GTR-O-SAT2_001.pdf
- Logan G, Freimanis GL, King DJ, Valdazo-González B, Bachanek-Bankowska K, Sanderson ND, et al. A universal protocol to generate consensus level genome sequences for foot-and-mouth disease virus and other positive-sense polyadenylated RNA viruses using the Illumina MiSeq. BMC Genomics. 2014;15:828. DOIPubMedGoogle Scholar
- Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:
vey016 . DOIPubMedGoogle Scholar - Templeton AR, Crandall KA, Sing CF. A cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data. III. Cladogram estimation. Genetics. 1992;132:619–33. DOIPubMedGoogle Scholar
- Callahan JD, Brown F, Osorio FA, Sur JH, Kramer E, Long GW, et al. Use of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth disease virus. J Am Vet Med Assoc. 2002;220:1636–42. DOIPubMedGoogle Scholar
- World Organisation for Animal Health. Foot and mouth disease (infection with foot and mouth disease virus). In: World Organisation for Animal Health, editor. Manual of diagnostic tests and vaccines for terrestrial animals. 2022 [cited 2024 Jan 10]. https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.01.08_FMD.pdf
- Gubbins S, Paton DJ, Dekker A, Ludi AB, Wilsden G, Browning CFJ, et al. Predicting cross-protection against foot-and-mouth disease virus strains by serology after vaccination. Front Vet Sci. 2022;9:
1027006 . DOIPubMedGoogle Scholar - McLaws M, Ahmadi BV, Condoleo R, Limon G, Kamata A, Arshed M, et al. Risk of foot-and-mouth disease SAT2 introduction and spread in countries in the Near East and West Eurasia–FAO qualitative risk assessment, October 2023. Rome: Food and Agriculture Organization of the United Nations; 2023. DOIGoogle Scholar
- Paton DJ, Sumption KJ, Charleston B. Options for control of foot-and-mouth disease: knowledge, capability and policy. Philos Trans R Soc Lond B Biol Sci. 2009;364:2657–67. DOIPubMedGoogle Scholar
- Di Nardo A, Knowles NJ, Paton DJ. Combining livestock trade patterns with phylogenetics to help understand the spread of foot and mouth disease in sub-Saharan Africa, the Middle East and Southeast Asia. Rev Sci Tech. 2011;30:63–85. DOIPubMedGoogle Scholar
- Food and Agriculture Organization of the United Nations. Report of the Seminar on the Control of Foot-and-Mouth Disease in the Near East. Ankara, Turkey; October 18–21, 1982 [cited 2024 Feb 26]. https://openknowledge.fao.org/items/4cbf774f-0782-4cd4-95b6-d86c10bf0f9e
- Ferris NP, Donaldson AI. The World Reference Laboratory for Foot and Mouth Disease: a review of thirty-three years of activity (1958-1991). Rev Sci Tech. 1992;11:657–84. DOIPubMedGoogle Scholar
- Canini L, Blaise-Boisseau S, Nardo AD, Shaw AE, Romey A, Relmy A, et al. Identification of diffusion routes of O/EA-3 topotype of foot-and-mouth disease virus in Africa and Western Asia between 1974 and 2019 - a phylogeographic analysis. Transbound Emerg Dis. 2022;69:e2230–9. DOIPubMedGoogle Scholar
- Al-Rawahi WA, Elshafie EI, Baqir S, Al-Ansari A, Wadsworth J, Hicks HM, et al. Detection of foot-and-mouth disease viruses from the A/AFRICA/G-I genotype in the Sultanate of Oman. Prev Vet Med. 2024;223:
106113 . DOIPubMedGoogle Scholar - The Pirbright Institute. FAO World Reference Laboratory for Foot-and-Mouth Disease Genotyping Report, Oman (WRLFMD). 2015 Jun 8 [cited 2024 Feb 26]. https://www.wrlfmd.org/sites/world/files/quick_media/WRLFMD-2015-00007-OMN-GTR-O-SAT2_001.pdf