Volume 31, Number 3—March 2025
Synopsis
Candida auris Outbreak and Epidemiologic Response in Burn Intensive Care Unit, Illinois, USA, 2021–2023
Cite This Article
Citation for Media
Abstract
Candida auris is an emerging fungal pathogen associated with outbreaks in healthcare settings. We report a multiyear outbreak of C. auris in a burn intensive care unit in Illinois, USA, during 2021–2023. We identified 28 C. auris cases in the unit over a 2-year period, despite outbreak response and multimodal mitigation measures. Of the 28 case-patients, 15 (53.6%) were considered colonized and 13 (46.4%) had clinical infections. Phylogenetic analysis of whole-genome sequences revealed 4 distinct clusters of closely related (0–6 SNP differences) genomes containing 3–6 cases. Clusters generally contained temporally related isolates from patients with epidemiologic links; this finding suggests that multiple introductions and within-unit spread over a limited time were responsible for the outbreak, rather than transmission from a long-term source (e.g., persistent environmental contamination or staff carriage). Here, integrated traditional and genomic epidemiology supported C. auris outbreak investigation and response and informed targeted interventions.
Candida auris is a fungal pathogen associated with colonization and high-mortality invasive infections in persons with underlying medical conditions, especially those who are hospitalized or reside in long-term care facilities (1,2). Prolonged skin colonization and environmental contamination likely contribute to within-facility persistence and spread (2–5). C. auris often displays extensive antifungal resistance and can acquire resistance rapidly during antifungal treatment (6–8).
Intensive care units (ICUs) are particularly vulnerable to C. auris outbreaks because of prolonged patient stays, high medical acuity, and extensive use of medical devices that can encourage pathogen spread (9–12). Effective infection prevention strategies are key to curbing the spread of C. auris; those strategies include contact screening, strict hand hygiene procedures, appropriate use of personal protective equipment (PPE) and transmission-based precaution by healthcare providers, use of single-patient equipment, environmental cleaning and disinfection, and private-room isolation (13). However, C. auris colonization and transmission have been reported to persist despite aggressive infection prevention interventions, making C. auris control a long-term burden in affected facilities (12,14,15).
In burn ICUs (BICUs), patients are at increased risk for healthcare-acquired infections because of breakdown of the skin barrier and the immunocompromising effects of burns; infection is the leading cause of death after burn injury (16). Fungal wound infections are reported in 6%–45% of all burn admissions; candidemia develops in up to 5% of patients with severe burns. Unlike most Candida species, C. auris has a tropism for skin (17), and it can readily colonize or infect adjacent large, open, nutrient-rich burn wounds. Furthermore, because they have frequent infections and large, open wounds, burn patients often require treatment with systemic and topical antimicrobials, both of which have capacity to eliminate competitive microbiota and encourage colonization with resistant organisms such as C. auris. Care provided in BICUs, such as skin debridement, may disperse colonized or infected skin cells into the environment, which contributes to transmission.
We describe a C. auris outbreak and response in a BICU in Illinois beginning in 2021. We used whole-genome sequencing (WGS) to help refine epidemiologic inferences and direct interventions. WGS has been used to support epidemiologic investigations of C. auris infection, including hospital outbreaks (9,18–23). Outbreak sequences generally form a unique clade with limited diversity (9,18); close relationships have been observed between epidemiologically linked cases (median 7 SNPs) and isolates from the same person (median 2 SNPs) (21). WGS can also detect antifungal resistance mutations (19,24,25). Thus, WGS may be a powerful tool to support C. auris outbreak investigations.
Study Setting and Participants
The Burn Center is a 10-bed intensive care unit caring for pediatric and adult burn patients at a 547-bed academic tertiary care medical center in the Chicago metropolitan area, Illinois, USA. The unit accommodates ICU overflow from other services, including medical and surgical ICUs. The unit practices universal contact precautions (gowns, gloves, masks, and eye protection) for all patients, staff, and visitors to the unit. We abstracted patient data via retrospective review of the hospital electronic medical records.
The Institutional Review Board of Loyola University (Chicago, IL, USA) reviewed and approved the protocol for this study (LU218571). Informed consent was waived.
Case Identification and Investigation
The outbreak investigation, led by the infection prevention team, consisted of admission screening and weekly point prevalence surveys of all patients in the unit. We defined a hospital-acquired case of C. auris as any illness in patient who, after a negative C. auris admission screen, tested positive for C. auris on subsequent weekly point prevalence screens or in any clinical specimen. We defined colonized cases as patients who had C. auris identified from surveillance cultures but no detection of C. auris in any clinical specimens. Clinical cultures refer to blood, wound, respiratory, or urine cultures.
We conducted epidemiologic investigations to identify commonalities between cases, including healthcare workers, medical equipment, prior room occupancies, and exposure locations outside of the BICU, including the operating room, tub room, and procedural areas such as the interventional radiology and gastroenterology suites. We reviewed patients’ history of C. auris through query of the Illinois extensively drug-resistant organism registry (34).
Infection Control Measures
Universal contact precautions and masks are used for all BICU patients; further containment strategies implemented in response to this outbreak involved increased observation of isolation compliance, education of nursing and ancillary staff about C. auris transmission and control, environmental cleaning validation, enhanced environmental cleaning with ultraviolet (UV) light, observation and training regarding correct use of PPE, and proper hand hygiene. In addition, the local health department performed an infection control assessment and response to identify and address infection control gaps.
Microbiologic Identification of Cases
We isolated C. auris fungus from screening samples collected from the axilla and groin of patients using a BBL CultureSwab EZ Collection and Transport System (BD, https://www.bd.com). We then inoculated samples onto HardyCHROM Candida (Hardy Diagnostics, https://hardydiagnostics.com) and incubated aerobically, protected from light, at 35°C for 72 hours. We isolated C. auris from clinical samples submitted for routine diagnostic testing on standard microbiologic media including sheep blood agar, chocolate agar, inhibitory mold agar (BBL prepared plated media; BD), and blood culture media (BACTEC Plus Aerobic and Lytic Anaerobic media; BD). We performed species identification by using Biotyper matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry with the MBT Compass Library version 12 Revision K (Bruker Daltonics, https://bruker.com).
Whole-Genome Sequencing
We suspended available C. auris isolates in DNA/RNAshield (Zymo, https://zymoresearch.com) and transported them to the Regional Innovative Public Health Laboratory at Rush University Medical Center (Chicago, IL, USA). We extracted nucleic acids using the Cultured Cells DNA Kit and Maxwell extraction system (Promega, https://promega.com) and prepared sequencing libraries using 1 ng DNA extract and Nextera XT DNA Library Preparation Kit (Illumina, https://illumina.com). We barcoded genome libraries by using IDT for Illumina DNA/RNA UD Indexes (Illumina) and balanced using a small-scale sequencing run of an equivolume pool (Illumina iSeq). We subjected final libraries to 2 × 150 paired-end sequencing on NovaSeq6000 (Illumina). We submitted data to the National Center for Biotechnology Information Short Read Archive (Appendix Table 1).
Bioinformatic and Statistical Analysis
We downloaded all publicly available Illinois C. auris sequences for comparison to outbreak sequences (Appendix Table 1). We analyzed paired-end sequences with the MycoSNP-nf pipeline version 1.4 (https://github.com/CDCgov/mycosnp-nf) by using clade IV reference B11243 (Genbank accession no. GCA_003014415.1) and implemented on Terra as previously described (20,27,28), excluding isolates with estimated coverage depth <25. We determined C. auris clade using phylogenetics with clade I–IV reference sequences. We used SNP differences between all samples and the reference to build a neighbor-joining tree using MEGA 11 (29) as previously described with 1,000 bootstrap replicates (21,30). To compare SNP differences, we used SNP distance matrices with all available Illinois sequences. To identify potential antifungal-resistance mutations, we used Snippy (31) to query for mutations in the FKS1, ERG11, TAC1b, MRR1, ERG3, and FUR1 genes. We compared mean SNP differences using Kruskal-Wallis nonparametric testing with adjusted significance between individual groups calculated using the Dunn multiple comparisons test.
Outbreak Investigation
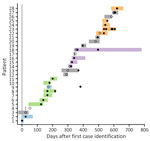
The first clinical C. auris isolate in a BICU patient was identified from blood culture in 2021. Three additional case-patients with hospital-acquired C. auris were identified in the subsequent 2 months (Figure 1). Admission screening in the BICU was initiated 3 months after the first case-patient was detected (Figure 1). A fifth case-patient, who originally screened negative on admission, was identified from a wound culture 83 days after admission; that case was notable because it was the first confirmed hospital acquisition of C. auris. A point prevalence survey 5 months after first case detection identified a sixth case. Weekly point prevalence screening was initiated 6 months after first case detection; 22 additional cases were identified 6–21 months after first case identification (Figure 1). Weekly point prevalence surveys were discontinued 28 days after discharge of the last patient with C. auris.
We reviewed case records to identify documented epidemiologic links; specifically, common locations (rooms), procedures, and staff exposures. Intensive observation of infection control practices throughout the BICU identified breaches that may have contributed to C. auris transmission, including poor hand hygiene compliance, improper PPE donning and doffing, cluttered patient care areas preventing thorough environmental cleaning, poor auditing of environmental cleaning, and inconsistent cleaning and disinfection practices for shared equipment. Shared equipment within the unit included bladder scanners, forced-air patient warming devices, vascular Dopplers, EKG machines, point-of-care ultrasounds, and recliners. Particular attention was given to staff whose patient care activities were extended to areas in the medical center outside of the BICU, including physical, occupational, speech and respiratory therapy, and radiology staff. The hospital infection control team and the local health department observed infection control breaches during the infection control assessment performed 6 months after the first case of C. auris was detected.
Outbreak Mitigation Measures
Early in the outbreak, a multidisciplinary C. auris response team, including staff from infection prevention, environmental services, nursing, facilities management, BICU physicians and hospital leadership, convened to create and implement a structured plan. All patients in the BICU with positive C. auris culture results were placed on contact precautions; signs were placed on the patients’ room doors, and their electronic medical records were flagged for C. auris and an isolation order. The team implemented outbreak mitigation measures universally in the BICU and centered on communication, education, and process improvement, focusing on environmental cleaning and hand hygiene. Education on C. auris transmission and necessary precautions were extended to the BICU nursing staff, with special attention on ancillary groups, particularly those also providing care to units outside of the BICU: environmental service, respiratory therapy, physical and occupational therapy, food and nutrition services, radiology, and pastoral care.
The team reviewed cleaning responsibilities between nursing and environmental service, including method of cleaning and frequency. Standard cleaning practices include floor and surface cleaning with a disinfectant effective against C. auris, bleach-wipe cleaning of equipment, and a log to track cleaning of shared equipment. Storage cabinets were installed in patient rooms. Black-light audits on discharge cleans were required on every terminal discharge to monitor cleaning practices, and environmental service staff received coaching when cleaning failures were identified. Germicidal ultraviolet disinfection was performed in patient rooms and above the unit’s nursing station beginning 9 months after first case detection. Thirteen months after first case detection, terminal cleaning of patient rooms incorporated high-intensity UV disinfection.
Unobtrusive-observer audits revealed that overall hand hygiene compliance was 78%–93% during the outbreak period. Most observations were of nursing staff; the greatest opportunities for improvement in compliance were among patient transporters (32% compliance), food and nutrition services (35% compliance), and physicians (67% compliance). The team increased hand hygiene promotion signage and efforts to normalize just-in-time coaching for hand hygiene and PPE breaches among staff and visitors.
Patient Characteristics
During the 21-month investigation, 28 patients were colonized or infected with C. auris (Table); 4 patients had invasive C. auris before admission screening. The average patient age was 49 years (range 16–81 years). Most patients were admitted with burns (64%), 9 patients (32%) were admitted with soft-tissue infections, and 1 patient was on medical ICU service. None of the patients had a history of C. auris infection, determined by chart review and query of the Illinois extensively drug-resistant organism registry. Seven patients were admitted from outside hospitals or had a hospitalization <30 days before the BICU admission; none were admitted from skilled nursing facilities. The mean length of stay in the BICU before identification of C. auris was 26 days (range 7–83 days). C. auris was identified in clinical cultures from 13 patients, some of which had C. auris in multiple cultures; 8 had C. auris identified in blood culture, 6 in respiratory culture, 8 in wound culture, and 3 in urine culture. The mean total length of stay in the BICU was 67 days (Figure 2).
Genomic Analysis of Outbreak Isolates
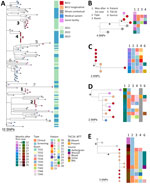
To investigate the genetic relationship of C. auris among cases, we conducted WGS on available isolates from the BICU outbreak, including isolates from 22 (79%) of 28 case-patients and 8 longitudinal isolates from 3 of those patients (Figure 2). Isolates from the remaining 6 case-patients were not available for analysis. We also sequenced available contemporaneous isolates from the same facility (n = 19) or another healthcare facility within the medical system (n = 31) to estimate diversity of hospital C. auris isolates and identify potential links outside of BICU. Of 80 isolates, 78 (97.5%) had sufficient genome quality for analysis; all were C. auris clade IV. Comparison with publicly available C. auris sequences from Illinois (n = 364) revealed that genomes from the BICU and related facilities were interspersed throughout other Illinois C. auris sequences (Appendix Figure 1), indicating multiple transmissions to the BICU facility from the broader diversity in the region.
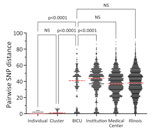
BICU isolates formed 4 clusters within Illinois sequences (Figure 3, panel A). Clusters contained 3–6 unique patients and closely related genomes (0–8 SNP differences) (Figure 3, panels B-E). Three BICU isolates did not cluster closely with any other BICU or Illinois isolate, differing by 7–39 SNPs from the closest non-BICU and 21–49 SNPs from the closest BICU isolate (Figure 3, panel A; Appendix Figure 1). We identified a mean 1.9 (range 0–8) SNP differences within BICU clusters, which was not significantly different from SNP differences from isolates collected from the same patient (p>0.9999) (Figure 4). However, SNP differences among all BICU isolates were significantly higher (mean 34.6; p<0.0001), but not significantly different from, mean SNP differences among isolates collected within the same timeframe elsewhere within the medical center (mean 35.8) or all Illinois (mean 37.9). Those findings indicate that BICU clusters were very closely related among isolates within the cluster but not more closely related among clusters than for other regional isolates, consistent with multiple independent introductions from regional C. auris followed by within-unit spread.
Integrated Genomic and Epidemiologic Investigation of Outbreak Clusters
Clusters 2 and 4 contained exclusively BICU isolates (Figure 3, panels C, E), whereas clusters 1 and 3 contained sequences collected within the medical center but outside of the BICU (Figure 3, panels B, D). In one instance, in cluster 1 (Figure 3, panel B), 1 sequence from the same facility collected 4 months before the first BICU case was identical to the first BICU case’s genome (patient 1). No epidemiologic links to BICU patients in this cluster were identified, and facility stays were separated by 123 days. In cluster 3 (Figure 3, panel D), 1 sequence from another unit within the facility and 1 sequence from elsewhere in the medical system collected 1 month before and 1 month after the first BICU case clustered with BICU isolates (patients 14, 15, 16, 18). No epidemiologic links were identified between the cases from the same facility. The case-patient from elsewhere in the medical system had been hospitalized at the BICU facility the month before, overlapping with other patients in this cluster. In addition, this patient and another BICU patient with C. auris from this cluster were both exposed to the same healthcare worker (speech therapist) during overlapping timeframes. Contextual isolates from institutions outside of the medical system did not fall in BICU clusters (>8 SNPs) (Figure 3; Appendix Figure 1).
BICU sequences clustered by collection date; clusters generally contained isolates collected within 3–6 months of each other (Figure 3, panels B–E). Clustering patients had overlapping BICU admission dates in all but 1 instance (Figure 2). In the exception, a patient (patient 9) from cluster 1 was admitted to the BICU 91 days after other patients in cluster 1 were discharged, but the C. auris isolate was not sequenced. This patient was then discharged to the same long-term acute care hospital as another patient in cluster 1 (Figure 3, panel B). The isolate belonging to cluster 1 was identified in an admission screening culture 8 months later, when the patient was admitted to another unit within the BICU facility. Thus, C. auris transmission may have occurred either on the BICU or at the long-term acute care hospital.
Analysis of Longitudinal Outbreak Isolates
Longitudinal isolates were collected from 3 patients over 51 (n = 2), 63 (n = 6), and 135 (n = 3) days and included both clinical and screening isolates. All longitudinal sequences clustered closely with other sequences from the same patient. Six of 11 longitudinal sequences were identical to another sequence from the same person; clinical and screening isolates were often identical to one another (Figure 3, panels C–E). Specimens collected from the same person had a mean 1.2 (range 1–4) SNP differences (Figure 4).
We investigated mutations in antifungal resistance–associated genes to look for longitudinal acquisition of antifungal resistance mutations. We identified a mutation associated with azole resistance in the TAC1b gene (I187T) in the last isolate collected from 1 patient; the 2 isolates obtained from this person earlier did not contain this mutation (Figure 3, panel D) (32,33). The patient received voriconazole therapy after collection of the isolates lacking the mutation but before the emergence of the TAC1b mutation. The isolate was not subjected to phenotypic antifungal susceptibility testing.
We describe a C. auris outbreak in a BICU that resulted in 28 patients colonized or infected over 2 years. We initially hypothesized that this was one continuous outbreak with possible environmental reservoirs on the unit contributing to ongoing transmission. WGS revealed 4 distinct clusters and 7 distinct genotypes; integration with epidemiologic information identified a complex outbreak that was driven both by importation of new strains and by within-BICU cross-transmission. Two phylogenetic clusters that included 7 (32%) of the sequenced case patient isolates contained isolates from both BICU patients and patients who were cared for in other units within the medical center, suggesting that C. auris might have been imported into the BICU from elsewhere in the facility by contaminated healthcare provider hands or clothing or by shared equipment. Alternatively, a BICU patient might have acquired C. auris upon exposure to contaminated surfaces or equipment in a common diagnostic or procedure area outside of the BICU. Either of those pathways could have led to the index BICU case; the index isolate genome was identical to an isolate collected from a patient in a unit elsewhere in the medical center 4 months earlier. However, epidemiologic links between new case-patients on the BICU and other patients in the hospital were not always identified.
Occult colonization, or colonization at low level or unsampled sites that results in nondetection by surveillance culture, of newly admitted patients might also have contributed to importation of C. auris into the BICU. Although all patients underwent axilla or groin screening for C. auris at the time of BICU admission, the sensitivity of this approach has been reported at ≈62%; to detect 100% of colonized patients, >6 body sites needed to be screened (15). Indeed, 3 patients were colonized with unique isolates that did not fall into any of the 4 clusters. Further, 11 of 22 case-patients whose isolates were sequenced and who had temporally overlapping BICU stays were included in 2 clusters that included only BICU patient isolates, suggesting within-BICU transmission of C. auris. Thus, undetected colonization at the time of admission and infection control breaches likely enabled introduction and transmission of C. auris to occur on the BICU. As our study demonstrated, C. auris is transmitted easily in healthcare facilities, and regional transmission can be hastened by patient transfers; in this outbreak, 61% of colonized or infected patients were discharged to other healthcare facilities. Interfacility communication and strict infection control measures are necessary to limit spread to other patient populations.
Once C. auris was introduced on the BICU, transmission was likely exacerbated by observed infection control breaches, particularly poor hand hygiene practices and lapses in cleaning of shared equipment. The prolonged lengths of stay of the patients (mean 67 days) also pose infection prevention and control challenges; C. auris rooms are recontaminated in as little as 4 hours after disinfection (34), emphasizing the need for stringent long-term adherence to cleaning and basic infection control practices.
The first limitation of our study is that it was conducted retrospectively; we selected samples for WGS on the basis of availability of stored isolates, and not all isolates from the outbreak were sequenced. Second, in most cases, only 1 isolate per patient was available for sequencing. Although some studies have found that patients can carry multiple genetically distinct C. auris isolates, genetically similar isolates may be more likely in an acute outbreak setting (9,21). In our study, all isolates collected from the same patient were closely related. Third, collection of epidemiologic metadata was limited to medical record review; some activities would not be recorded in the medical record. Further, contamination of portable unit-based equipment that is shared between patients might have contributed to ongoing C. auris transmission, but this possibility could not be verified through medical record review, and we conducted no environmental culturing.
WGS refined our understanding of this C. auris outbreak. The discovery that the outbreak included multiple introductions of C. auris onto the unit influenced our current approach to C. auris investigation and response; we focus now on between-unit transmission, including the possible role of ancillary personnel who move throughout the hospital, and not just on within-unit infection prevention measures. Furthermore, we conduct admission screening as well as point prevalence survey protocols in response to a C. auris case to identify and isolate colonized patients quickly. Integrated WGS and epidemiologic investigation is a powerful tool for identifying drivers of transmission in nosocomial outbreaks.
Dr. Barbian is an assistant professor and genomic epidemiologist at Rush University Medical Center Regional Innovative Public Health Laboratory. Her primary research interests are public health genomics, genomic surveillance, and pathogen evolution.
Acknowledgments
We thank Stefan Green, Sofiya Bobrovska, and members of the Rush University Medical Center Genomics and Microbiome Core Facility for whole-genome sequencing and data management. We thank Michael Bauer and members of the Loyola University Medical Center clinical microbiology laboratory for generation, storage, and transport of isolates. We thank Isaac Ghinai and the Chicago Department of Public Health Laboratory-Based Surveillance and Healthcare Settings teams. We thank Robert Garcia for manuscript review.
This project was supported by the Centers for Disease Control and Prevention (CDC) of the US Department of Health and Human Services (HHS) as part of a financial assistance award totaling $11,162,000 with 100% funded by CDC/HHS. The contents are those of the authors and do not necessarily represent the official views of, nor an endorsement by, CDC/HHS or the US government.
References
- Tsay S, Welsh RM, Adams EH, Chow NA, Gade L, Berkow EL, et al.; MSD. Notes from the field: ongoing transmission of Candida auris in health care facilities—United States, June 2016–May 2017. MMWR Morb Mortal Wkly Rep. 2017;66:514–5. DOIPubMedGoogle Scholar
- Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, et al. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus-United States, May 2013–August 2016. Am J Transplant. 2017;17:296–9. DOIPubMedGoogle Scholar
- Piedrahita CT, Cadnum JL, Jencson AL, Shaikh AA, Ghannoum MA, Donskey CJ. Environmental surfaces in healthcare facilities are a potential source for transmission of Candida auris and other Candida species. Infect Control Hosp Epidemiol. 2017;38:1107–9. DOIPubMedGoogle Scholar
- Sexton DJ, Bentz ML, Welsh RM, Derado G, Furin W, Rose LJ, et al. Positive correlation between Candida auris skin-colonization burden and environmental contamination at a ventilator-capable skilled nursing facility in Chicago. Clin Infect Dis. 2021;73:1142–8. DOIPubMedGoogle Scholar
- Horton MV, Johnson CJ, Kernien JF, Patel TD, Lam BC, Cheong JZA, et al. Candida auris forms high-burden biofilms in skin niche conditions and on porcine skin. MSphere. 2020;5:e00910–9. DOIPubMedGoogle Scholar
- Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017;64:134–40. DOIPubMedGoogle Scholar
- Biagi MJ, Wiederhold NP, Gibas C, Wickes BL, Lozano V, Bleasdale SC, et al. Development of high-level echinocandin resistance in a patient with recurrent Candida auris candidemia secondary to chronic candiduria. Open Forum Infect Dis. 2019;6:
ofz262 . DOIPubMedGoogle Scholar - Rybak JM, Barker KS, Muñoz JF, Parker JE, Ahmad S, Mokaddas E, et al. In vivo emergence of high-level resistance during treatment reveals the first identified mechanism of amphotericin B resistance in Candida auris. Clin Microbiol Infect. 2022;28:838–43. DOIPubMedGoogle Scholar
- Eyre DW, Sheppard AE, Madder H, Moir I, Moroney R, Quan TP, et al. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018;379:1322–31. DOIPubMedGoogle Scholar
- Ruiz-Gaitán A, Moret AM, Tasias-Pitarch M, Aleixandre-López AI, Martínez-Morel H, Calabuig E, et al. An outbreak due to Candida auris with prolonged colonisation and candidaemia in a tertiary care European hospital. Mycoses. 2018;61:498–505. DOIPubMedGoogle Scholar
- Armstrong PA, Rivera SM, Escandon P, Caceres DH, Chow N, Stuckey MJ, et al. Hospital-associated multicenter outbreak of emerging fungus Candida auris, Colombia, 2016. Emerg Infect Dis. 2019;25:1339–46. DOIPubMedGoogle Scholar
- Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, et al. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016;5:35. DOIPubMedGoogle Scholar
- Jeffery-Smith A, Taori SK, Schelenz S, Jeffery K, Johnson EM, Borman A, et al.; Candida auris Incident Management Team. Candida auris Incident Management Team. Candida auris: a review of the literature. Clin Microbiol Rev. 2017;31:e00029–17.PubMedGoogle Scholar
- Pacilli M, Kerins JL, Clegg WJ, Walblay KA, Adil H, Kemble SK, et al. Regional emergence of Candida auris in Chicago and lessons learned from intensive follow-up at 1 ventilator-capable skilled nursing facility. Clin Infect Dis. 2020;71:e718–25. DOIPubMedGoogle Scholar
- Proctor DM, Dangana T, Sexton DJ, Fukuda C, Yelin RD, Stanley M, et al.; NISC Comparative Sequencing Program. Integrated genomic, epidemiologic investigation of Candida auris skin colonization in a skilled nursing facility. Nat Med. 2021;27:1401–9. DOIPubMedGoogle Scholar
- Escandón-Vargas K, Tangua AR, Medina P, Zorrilla-Vaca A, Briceño E, Clavijo-Martínez T, et al. Healthcare-associated infections in burn patients: Timeline and risk factors. Burns. 2020;46:1775–86. DOIPubMedGoogle Scholar
- Huang X, Hurabielle C, Drummond RA, Bouladoux N, Desai JV, Sim CK, et al. Murine model of colonization with fungal pathogen Candida auris to explore skin tropism, host risk factors and therapeutic strategies. Cell Host Microbe. 2021;29:210–221.e6. DOIPubMedGoogle Scholar
- Eckbo EJ, Wong T, Bharat A, Cameron-Lane M, Hoang L, Dawar M, et al. First reported outbreak of the emerging pathogen Candida auris in Canada. Am J Infect Control. 2021;49:804–7. DOIPubMedGoogle Scholar
- Rhodes J, Abdolrasouli A, Farrer RA, Cuomo CA, Aanensen DM, Armstrong-James D, et al. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect. 2018;7:1–12. DOIGoogle Scholar
- Gorzalski A, Ambrosio FJ III, Massic L, Scribner MR, Siao DD, Hua C, et al. The use of whole-genome sequencing and development of bioinformatics to monitor overlapping outbreaks of Candida auris in southern Nevada. Front Public Health. 2023;11:
1198189 . DOIPubMedGoogle Scholar - Chow NA, Gade L, Tsay SV, Forsberg K, Greenko JA, Southwick KL, et al.; US Candida auris Investigation Team. Multiple introductions and subsequent transmission of multidrug-resistant Candida auris in the USA: a molecular epidemiological survey. Lancet Infect Dis. 2018;18:1377–84. DOIPubMedGoogle Scholar
- Chow NA, Muñoz JF, Gade L, Berkow EL, Li X, Welsh RM, et al. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. MBio. 2020;11:e03364–19. DOIPubMedGoogle Scholar
- Escandón P, Chow NA, Caceres DH, Gade L, Berkow EL, Armstrong P, et al. Molecular epidemiology of Candida auris in Colombia reveals a highly related, countrywide colonization with regional patterns in amphotericin B resistance. Clin Infect Dis. 2019;68:15–21. DOIPubMedGoogle Scholar
- Naicker SD, Maphanga TG, Chow NA, Allam M, Kwenda S, Ismail A, et al. Clade distribution of Candida auris in South Africa using whole genome sequencing of clinical and environmental isolates. Emerg Microbes Infect. 2021;10:1300–8. DOIPubMedGoogle Scholar
- Misas E, Escandón PL, Gade L, Caceres DH, Hurst S, Le N, et al. Genomic epidemiology and antifungal-resistant characterization of Candida auris, Colombia, 2016-2021. MSphere. 2024;9:
e0057723 . DOIPubMedGoogle Scholar - Trick WE, Lin MY, Cheng-Leidig R, Driscoll M, Tang AS, Gao W, et al. Electronic public health registry of extensively drug-resistant organisms, Illinois, USA. Emerg Infect Dis. 2015;21:1725–32. DOIPubMedGoogle Scholar
- Libuit KG, Doughty EL, Otieno JR, Ambrosio F, Kapsak CJ, Smith EA, et al. Accelerating bioinformatics implementation in public health. Microb Genom. 2023;9:
mgen001051 . DOIPubMedGoogle Scholar - Bagal UR, Phan J, Welsh RM, Misas E, Wagner D, Gade L, et al. MycoSNP: a portable workflow for performing whole-genome sequencing analysis of Candida auris. Methods Mol Biol. 2022;2517:215–28. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38:3022–7. DOIPubMedGoogle Scholar
- Welsh RM, Misas E, Forsberg K, Lyman M, Chow NA. Candida auris whole-genome sequence benchmark dataset for phylogenomic pipelines. J Fungi (Basel). 2021;7:214. DOIPubMedGoogle Scholar
- Seemann T. Snippy: rapid haploid variant calling and core genome alignment. Github [cited 2025 Jan 29]. https://github.com/tseemann/snippy
- Rybak JM, Muñoz JF, Barker KS, Parker JE, Esquivel BD, Berkow EL, et al. Mutations in TAC1B: a novel genetic determinant of clinical fluconazole resistance in Candida auris. MBio. 2020;11:e00365–20. DOIPubMedGoogle Scholar
- Carolus H, Pierson S, Muñoz JF, Subotić A, Cruz RB, Cuomo CA, et al. Genome-wide analysis of experimentally evolved Candida auris reveals multiple novel mechanisms of multidrug resistance. MBio. 2021;12:e03333–20. DOIPubMedGoogle Scholar
- Sansom SE, Gussin GM, Schoeny M, Singh RD, Adil H, Bell P, et al. Rapid environmental contamination with Candida auris and multidrug-resistant bacterial pathogens near colonized patients. Clin Infect Dis. 2024; 78:1276–84.
Figures
Table
Cite This ArticleOriginal Publication Date: February 21, 2025
1These first authors contributed equally to this article.
2These authors contributed equally to this article.
Table of Contents – Volume 31, Number 3—March 2025
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Hannah Barbian, Rush University Medical Center, Jelke 1412, 1750 W Harrison St, Chicago, IL 60612, USA
Top